Whole genome alignments, demo
Let’s look at some of the comparative genomics views in the Location tab. Go to the region 9:1928000-2017000 in zebrafish, which contains the hoxD cluster which is involved in limb development and is highly conserved between species.
We can look at pairwise alignment tracks in this view by clicking on Configure this page.
Select BLASTz/LASTz alignments from the left-hand menu to choose alignments between closely related species. Turn on the alignments for Chinook Salmon, Midas cichlid _ and _Sailfin molly in Normal. Save and close the menu.

The alignment is greatest between closely related species.
We can also look at the alignment between species or groups of species as text. Click on Alignments (text) in the left hand menu.
Select Select an alignment to open the alignment menu.
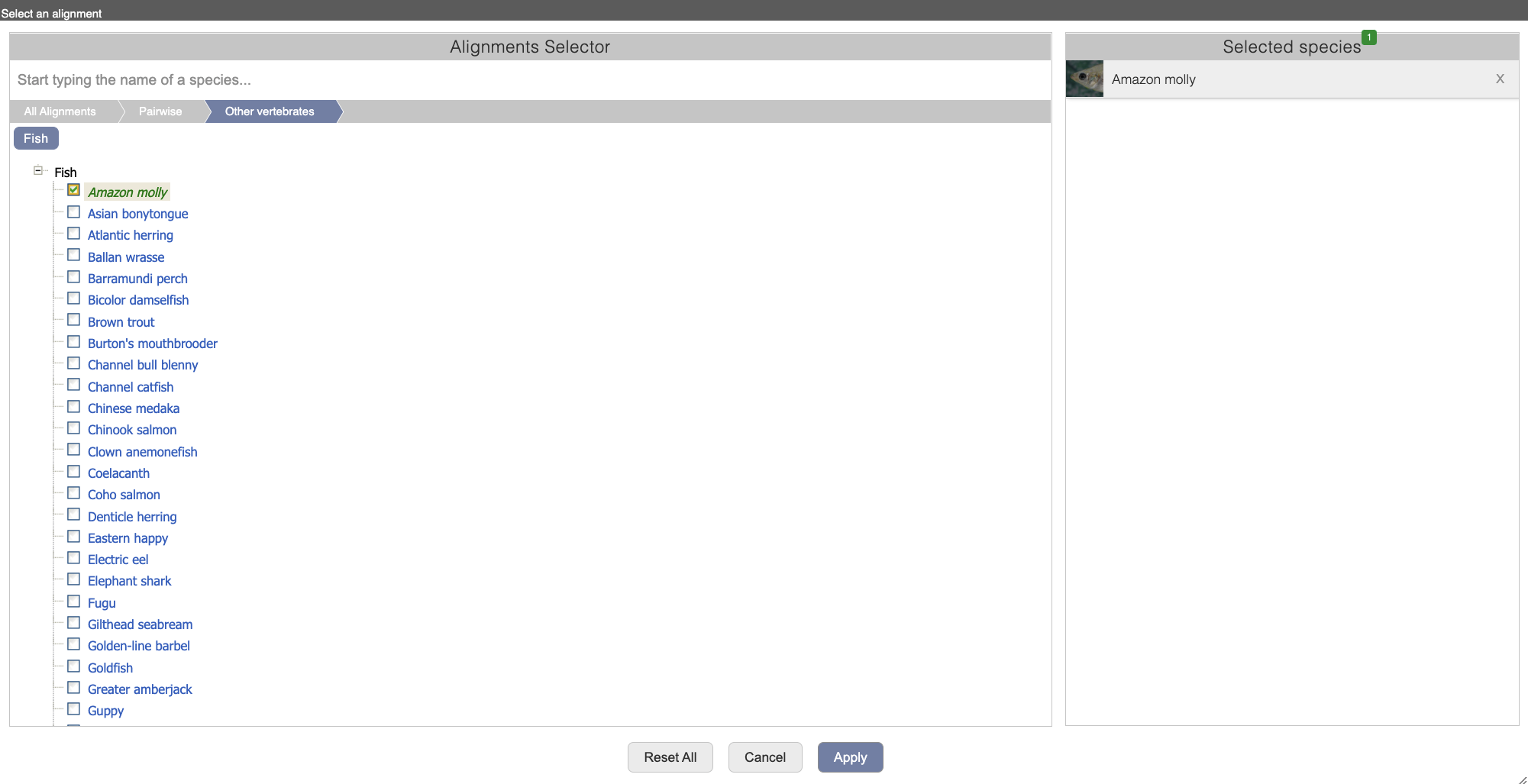
Click through the links, Pairwise, Other vertebrates, Fish to select Amazon molly.
In this case there are nine blocks aligned, Block 1 a large (73775 bp) alignment against Amazon molly scaffold KI519626.1 and four smaller blocks. Click on Block 1.
You will see a list of the regions aligned, followed by the sequence alignment. Exons are shown in red.
To compare with both contigs visually, go to Region comparison.
To add species to this view, click on the blue Select species or regions button. Choose Amazon molly again then close the menu.
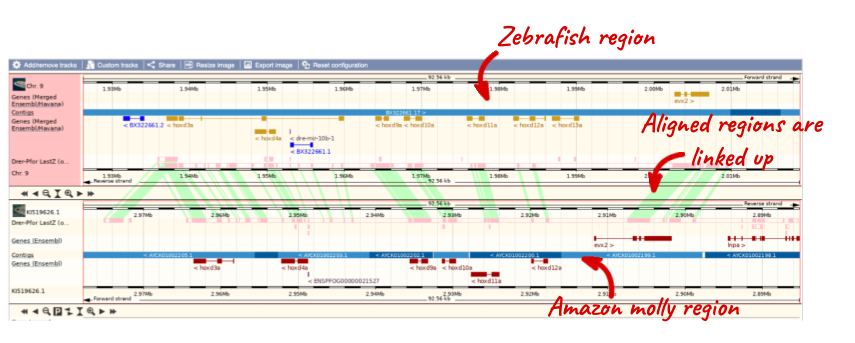
You can configure this view for both species. Click on Configure this page and look in the top left of the menu.

The drop down allows you to configure each species separately.
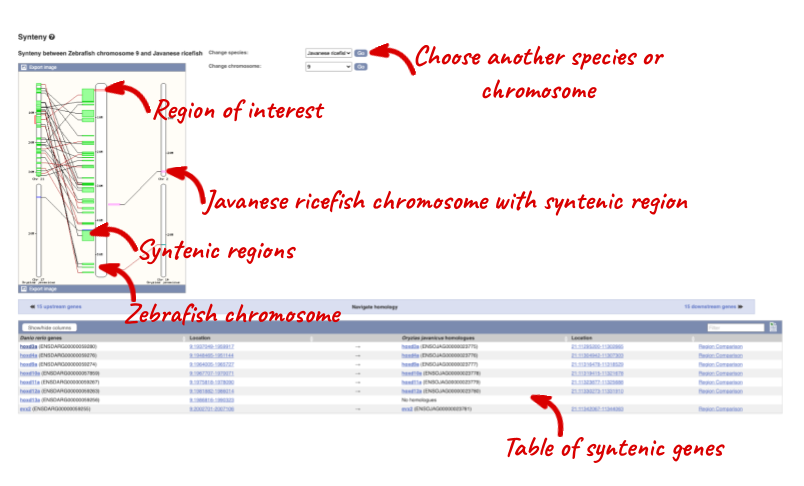
We can view large scale syntenic regions from our chromosome of interest. Click on Synteny in the left hand menu and select Javanese ricefish.