Finding information about species and genomes in Ensembl, Demo
Demo: Introduction to Ensembl
Ensembl
Homepage
The front page of Ensembl is found at ensembl.org. It contains lots of information and links to help you navigate Ensembl:
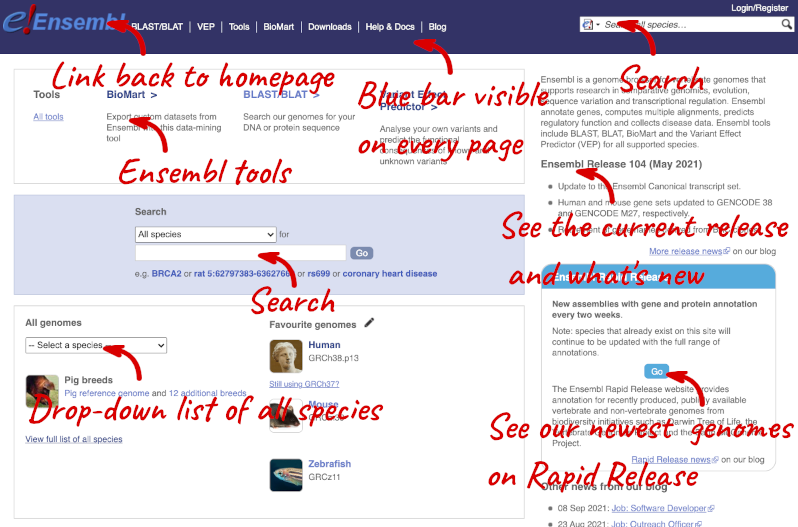
On the right-hand panel you can see the current release number and what has come out in this release. To access old releases, scroll to the bottom of the page and click on View in archive site in the right-hand corner.
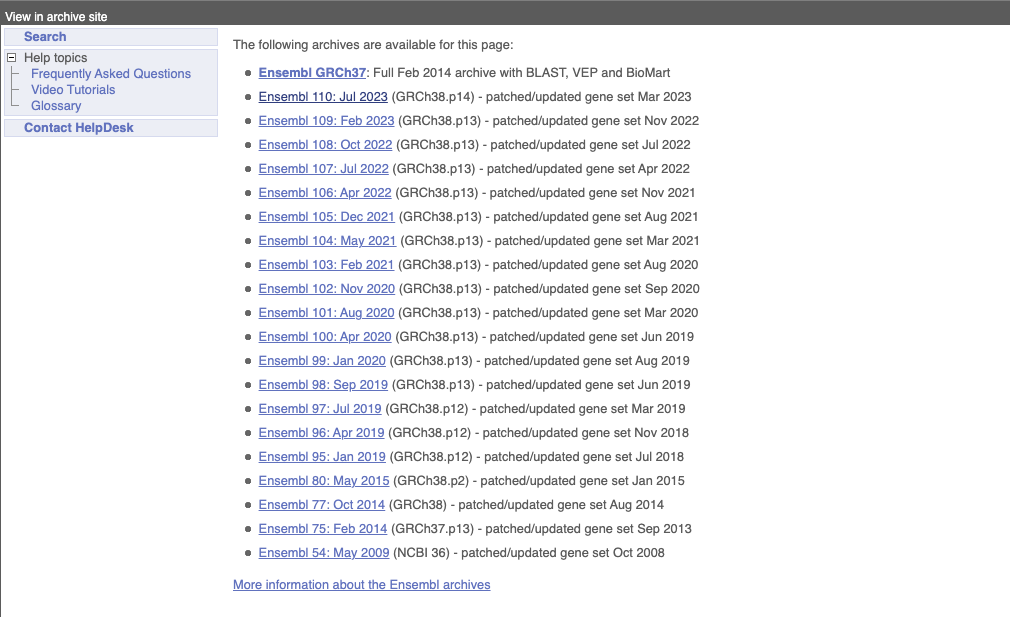
Click on the links to go to the archives. Alternatively, you can jump quickly to the correct release by adding e plus the release number in the URL. For example e98.ensembl.org jumps to Ensembl release 98.
Available species
Scroll back up to the top of the homepage. You can view all available species by clicking the View full list of all species link underneath the coloured search block.
You can search for your species of interest (either the common or scientific name) using the search bar at the top right-hand corner of the table. Click on the common name of your species of interest to go to the species information page. We’ll click on Human.
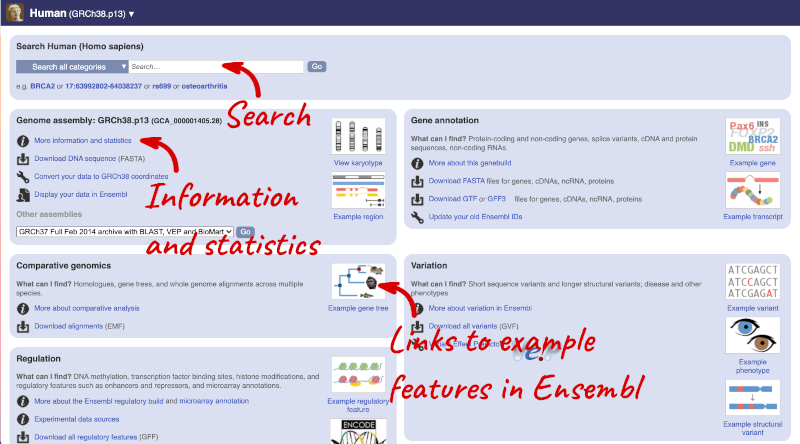
Species information
Here you can see links to example features and to download flatfiles. To find out more about the genome assembly and genebuild, click on More information and statistics under the Genome assembly section.
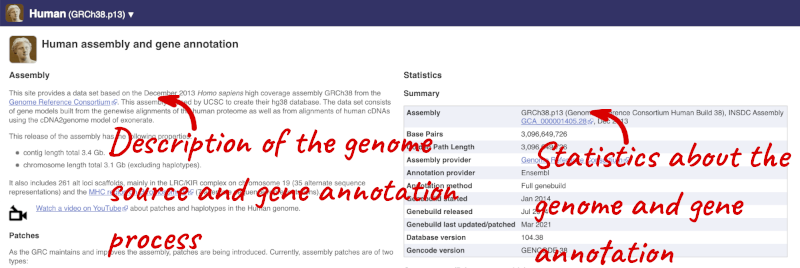
Here you’ll find a detailed description of how to the genome was produced and links to the original source. You will also see details of how the genes were annotated.
The current genome assembly for human is GRCh38. If you want to see the previous assembly, GRCh37, visit our dedicated site grch37.ensembl.org.
Ensembl Genomes
Homepage
Let’s take a look at the Ensembl Genomes homepage at ensemblgenomes.org.
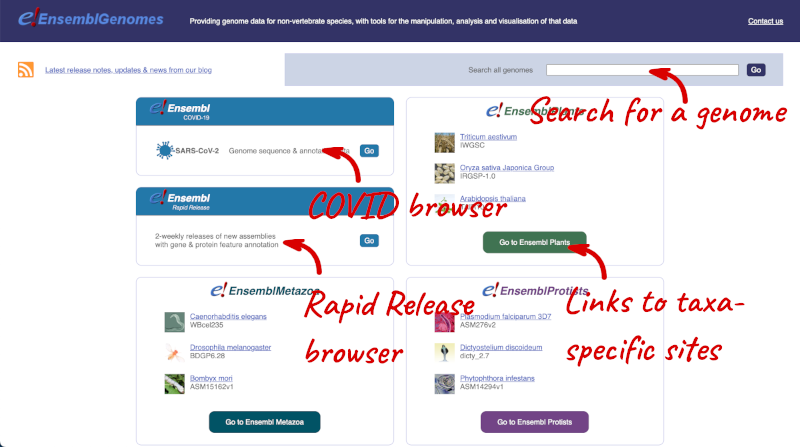
Click on the different taxa to see their homepages. Each one has a different colour-coding, but they are all structured in a similar format to the Ensembl main site.
You can navigate most of the taxa in the same way as you would with Ensembl.
Ensembl Bacteria
Ensembl Bacteria has a large number of genomes and has a slightly different method to the other Ensembl sites. Let’s look at it in more detail.
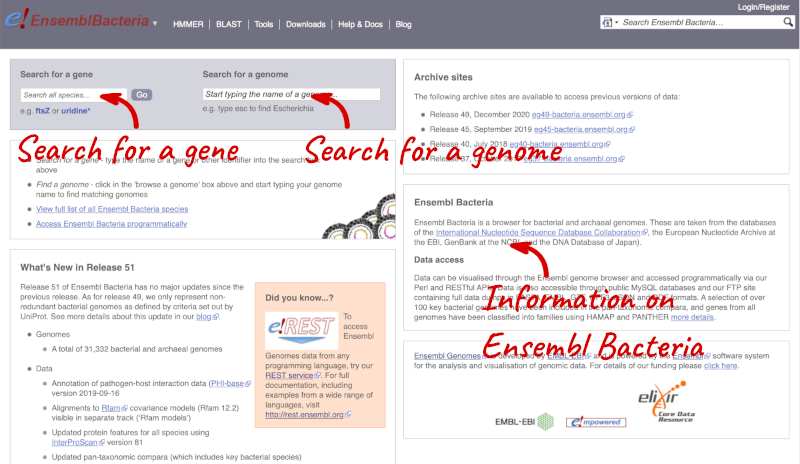
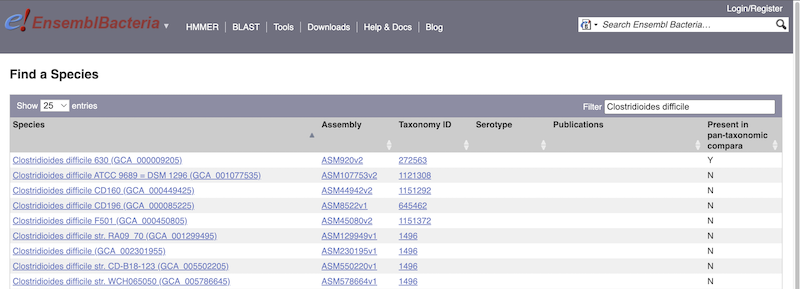
There’s no drop-down species list for bacteria as it would be hard to navigate with the number of species. You can click the View full list of all Ensembl Bacteria species link underneath the coloured search block. Search for your species of interest using the filter in the top right-hand corner of the table.
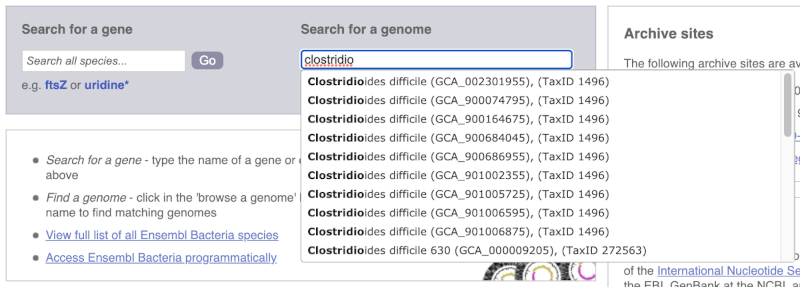
Alternatively, you can find a species by typing the species name into the Search for a genome search box at the top of the page. A drop-down list will appear with any species matching the name you entered.
For example, to find a sub-strain of Clostridioides difficile start typing in the species name. Due to the auto-complete, you’ll see useful results as soon as you get to Clostridio.
The drop down contains various strains of C. difficile. Let’s choose C. difficile 630. This will take us to another species information page, where we can explore various features.
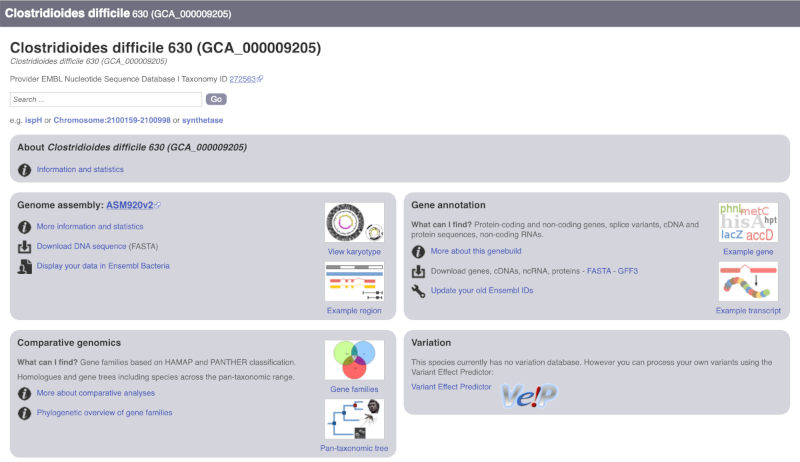
Unlike the Homo sapiens species information page, there is no prose description of the genome or gene annotation, as these pages were generated automatically.
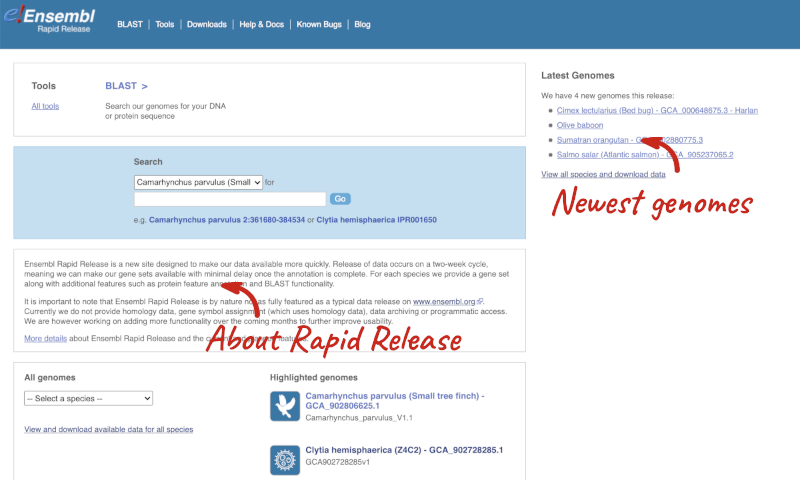
Ensembl Rapid Release
Our newest genomes, such as those coming from the Darwin Tree of Life, are available rapid.ensembl.org with limited annotation.