Gene trees and homologues, demo
Demo: gene trees and homology predictions
Comparative Genomics
Gene trees
Let’s look at the homologues of the Salmo salar (Atlantic salmon) gh1 (ENSSSAG00000055013) gene. This gene codes for growth hormone 1. Search for the gene and go to the Gene tab.
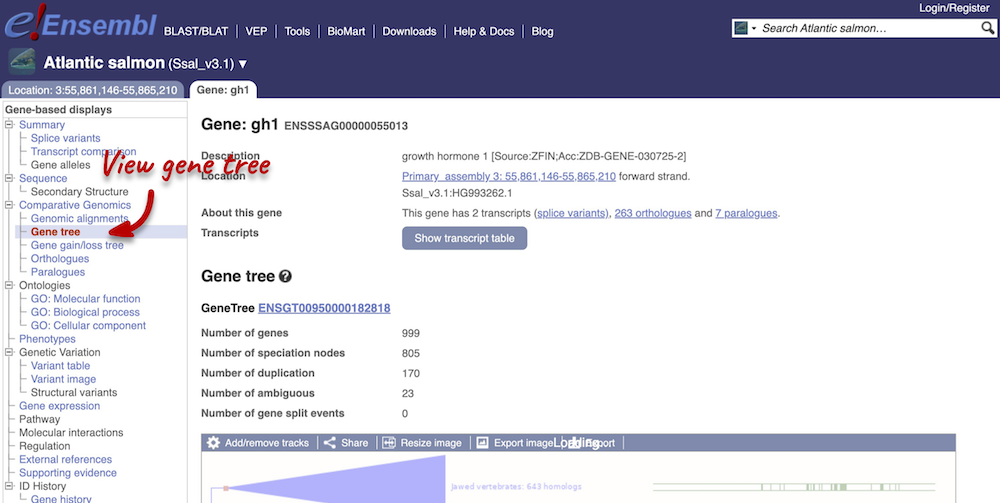
Click on Comparative Genomics: Gene tree. This will display the current gene in the context of a phylogenetic tree used to determine orthologues and paralogues.
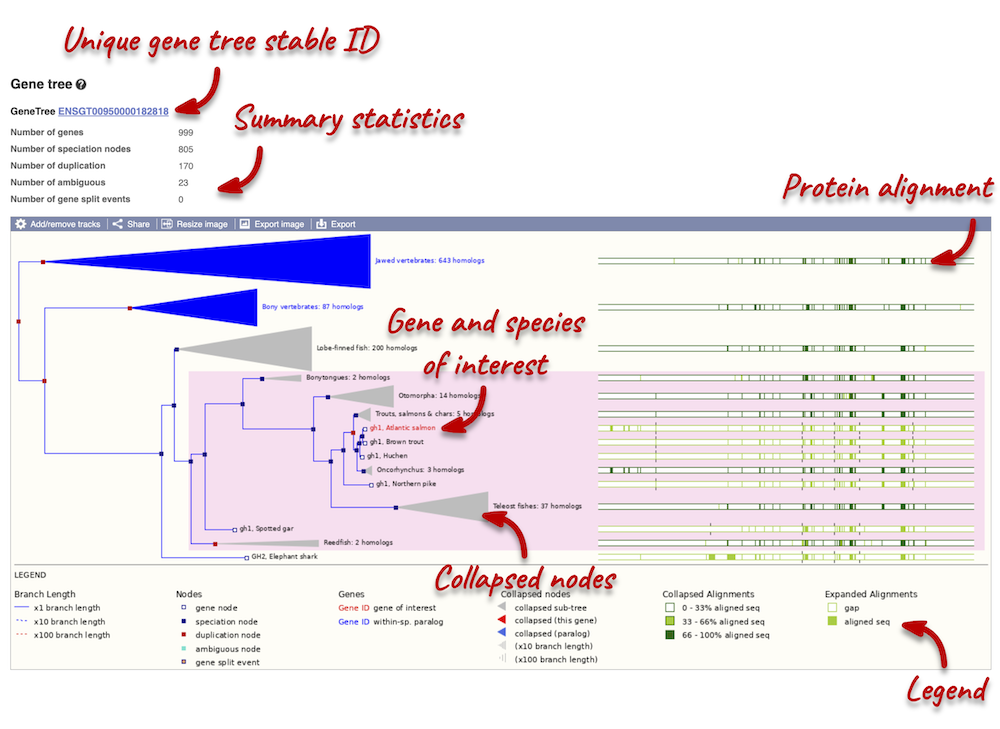
The phylogeny is displayed on the left-hand side. Protein alignments are displayed on the right-hand side. You can find a legend at the bottom of the view.
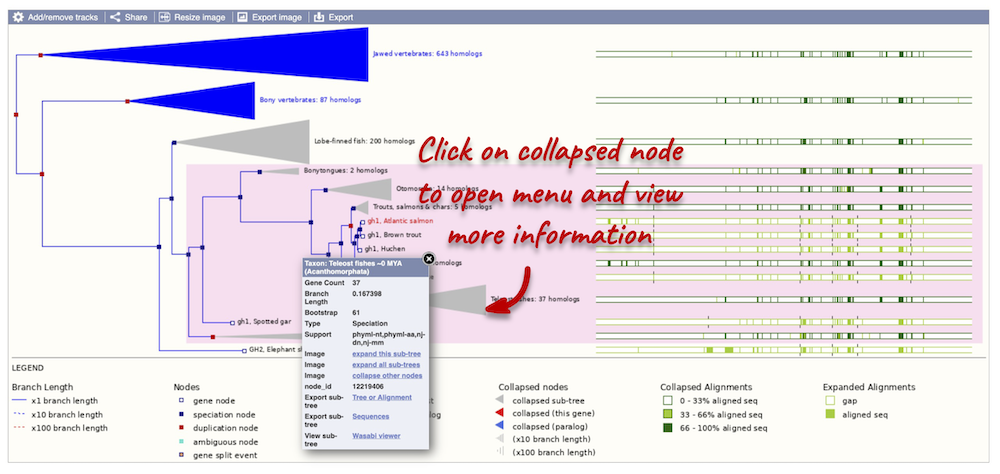
Funnels within the phylogeny indicate collapsed nodes. Click on a node (coloured triangle) to open a menu. We can then see what type of node this is, some statistics and options to expand or export the sub-tree.
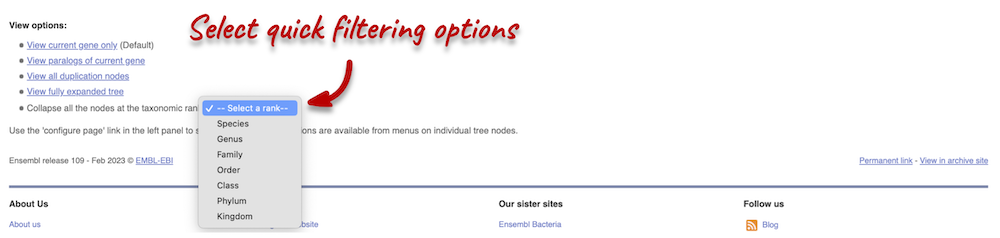
There are some quick filtering options below the image, where you can add paralogues, and quickly expand or collapse nodes.
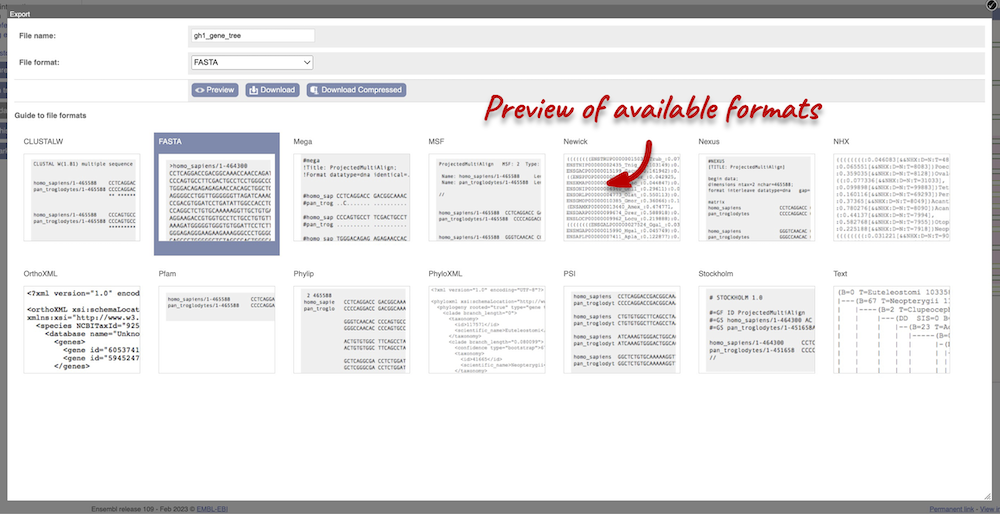
You can download the tree in a variety of formats. From the pop-up above, you can click to export the sub-tree (everything to the right of the node). Alternatively, click on the Export icon ![]() in the bar at the top of the image to get a pop-up where you can choose your format. You can preview this file before you download.
in the bar at the top of the image to get a pop-up where you can choose your format. You can preview this file before you download.
Homologues
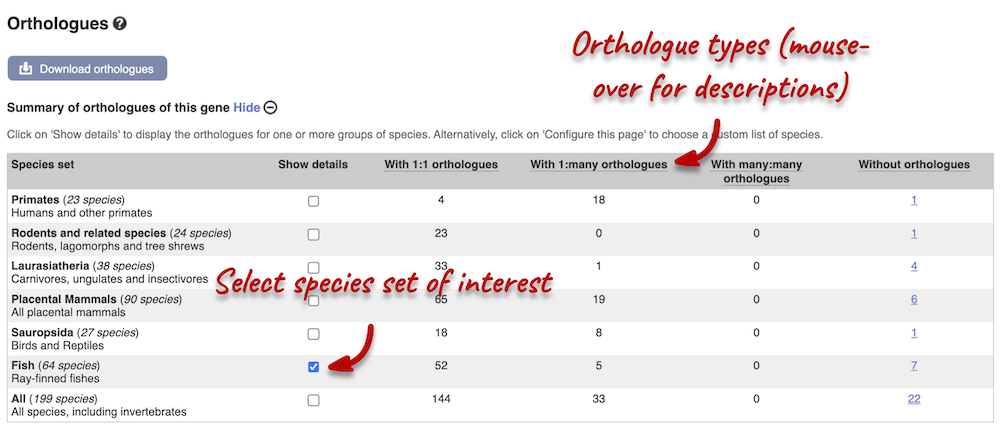
We can look at homologues in the Orthologues and Paralogues pages, which can be accessed from the left-hand menu. If there are no orthologues or paralogues, then the name will be greyed out. Click on Comparative Genomics: Orthologues to see the orthologues available. In the first table, you will find a summary of orthologues by species sets. Let’s select the Fish set only. There are 64 species in this set.
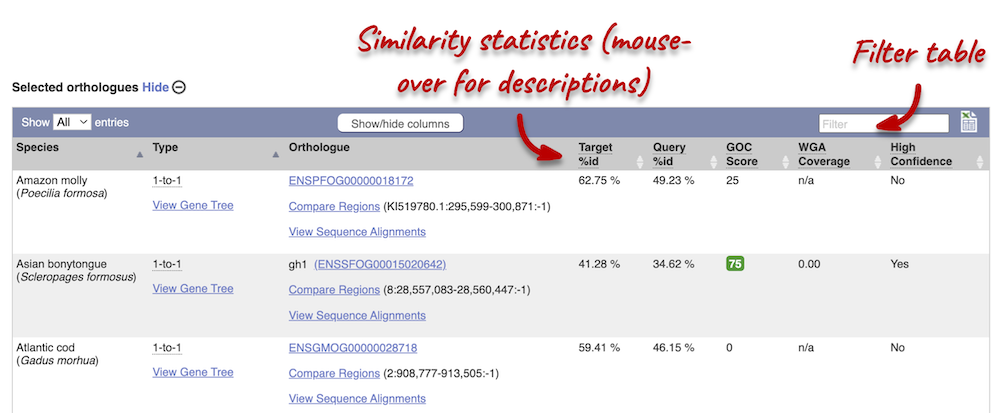
In the second table, you will find orthologue details per species:
Scroll to the bottom of the page to see a list of the species that do not have any orthologues with gh1 in Atlantic salmon… there are a lot!

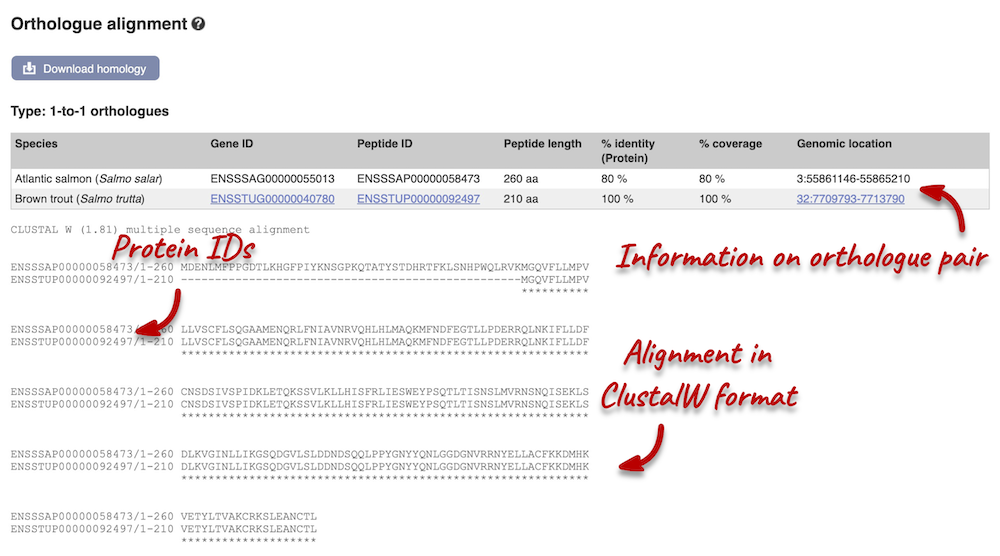
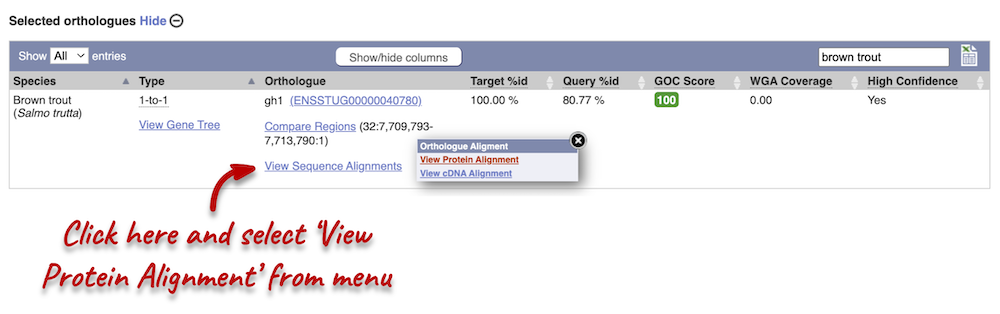
Let’s compare the Atlantic salmon and Salmo trutta (brown trout) homologues. Search for brown trout in the Orthologues table above using the filter at the top right-hand corner. Links from the orthologue allow you to view the two orthologues within a gene tree, see the gene entry for brown trout, compare the regions of the orthologues, or view protein and cDNA alignments. Click on View Sequence Alignments, and then View Protein Alignment for the brown trout orthologue.
The protein alignment is displayed in ClustalW format. If you are unfamiliar with this format, you can find information in the Ensembl help pages. You can also export the alignment in other formats by clicking the Download homology button at the top of the alignment.