Exporting gene data with BioMart, demo
Demo: BioMart
You have 4 Salmo salar (Atlantic salmon) genes:
slc45a2, igf1, sod1, ube2f
Follow these instructions to guide you through BioMart to answer the following questions:
- What are the NCBI Gene IDs for these genes?
- Are there associated functions from the Gene Ontology (GO) project that might help describe their function?
- What are the coordinates of the genes?
- What are their cDNA sequences?
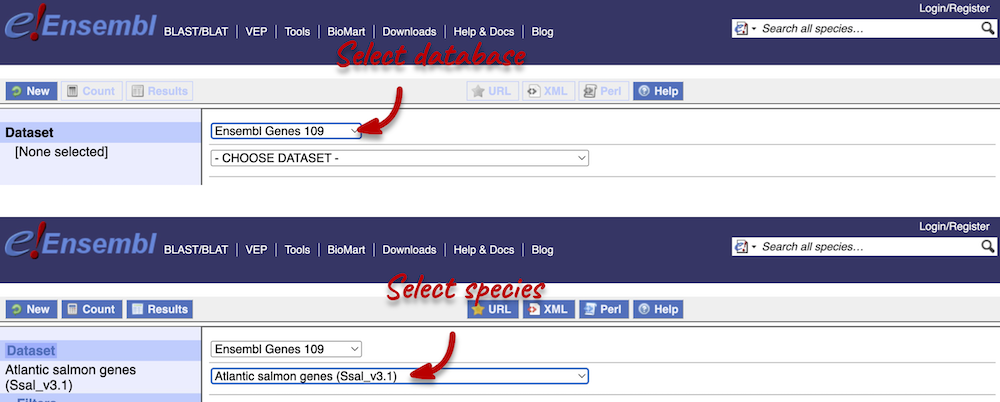
Step 1: Choose database and dataset
Click on BioMart located on the navigation bar at the top of any Ensembl page. Select Database: Ensembl Fungi Genes and Dataset: Atlantic salmon genes.
Step 2: Choose appropriate filters
We want to find these 4 genes in Atlantic salmon:
slc45a2, igf1, sod1, ube2f
We need to filter the dataset to look only at these genes. Click on Filter on the left-hand panel and open the GENES tab on the right.
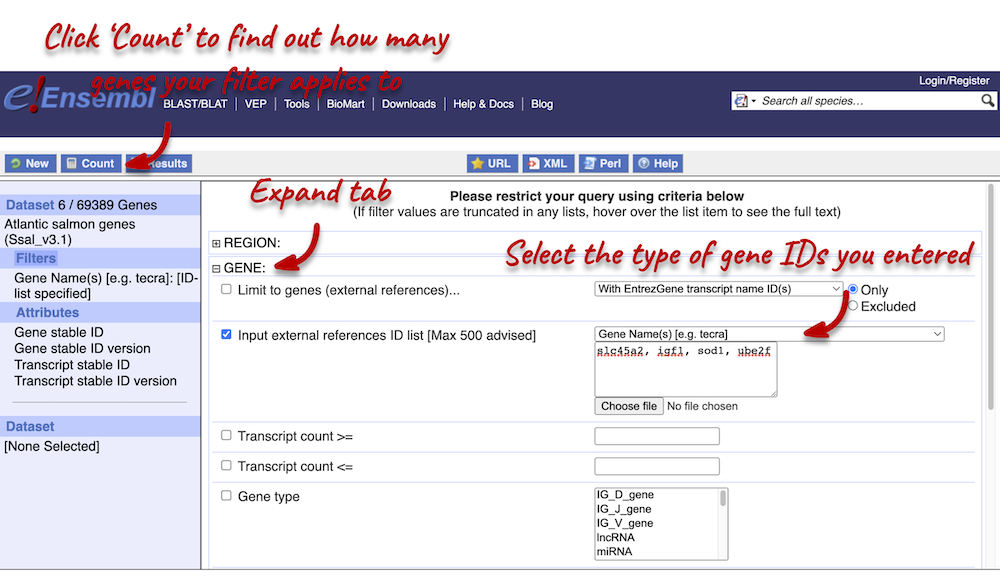
Using the count function we can see that our filter applies to 6 out of 69,389 genes in Atlantic salmon. The count is 6 (rather than 4) because gene names can be ambiguous. This means that a gene name can be given to multiple different genes.
Step 3.1: Select attributes (features)
Attributes are defined by what we would like to learn about the data. We want to find out more information about the genes’:
- NCBI IDs
- Associated GO terms
- Coordinates
- cDNA sequences
You can only select one attribute category at a time. We can answer points 1 and 3 in a single query, but we will need to do a second query to answer point 4. Make sure that Features category is selected at the top of the page. Expand the GENE tab and select the following features:
- Gene name
- Gene start
- Gene end
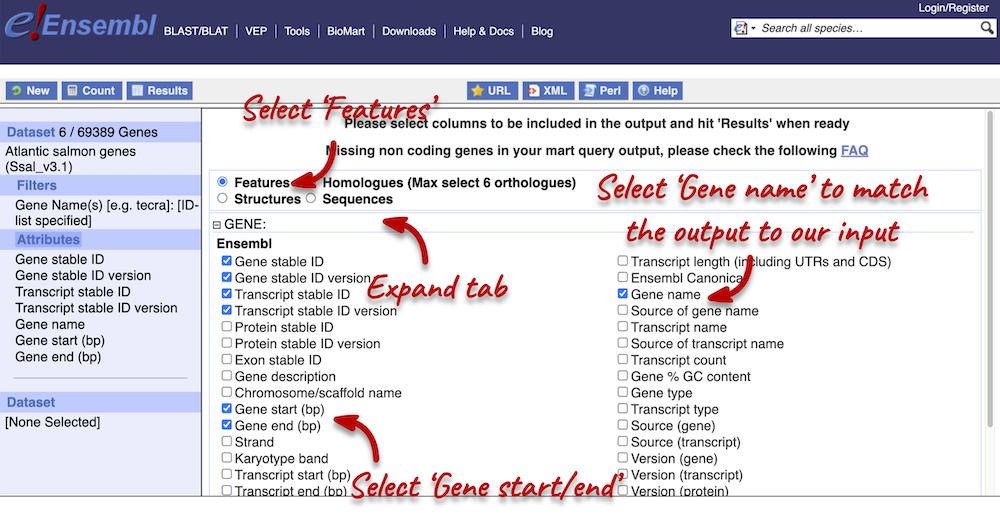
We have added the Gene name attribute, because we want to be able to match out output with our input. As we searched for gene names, we want to be able to see which features match to our given gene.
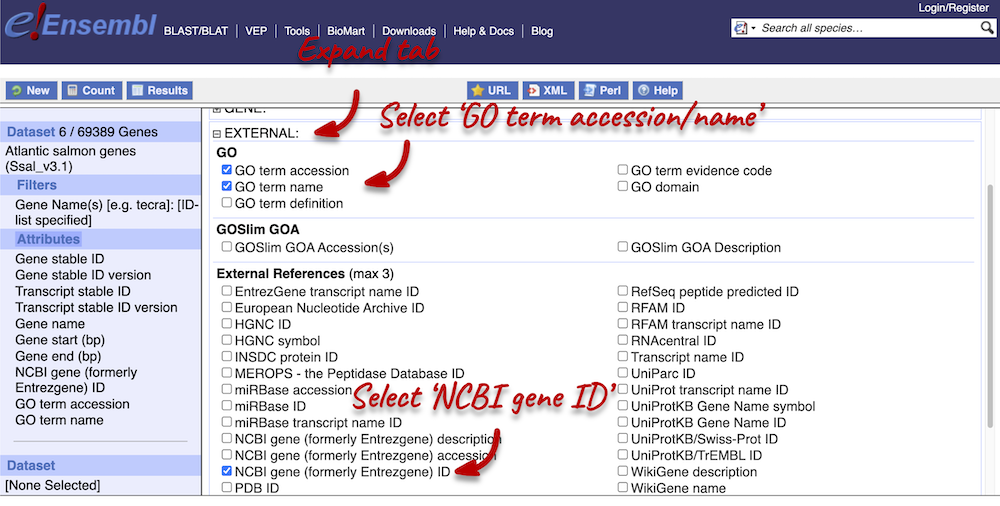
Next, expand the EXTERNAL tab. This section contains lots of identifiers from databases outside of Ensembl. Select the following features:
- NCBI IDs
- GO term accession
- GO term name
Step 4.1: Get the results (features)
You can download the data by selecting a format type (HTML, TSV, CSV or XLS) from the drop-down under Export all results to and clicking Go. The table presented shows a sub-sample of 10 results to enable you to check you have the correct attributes.
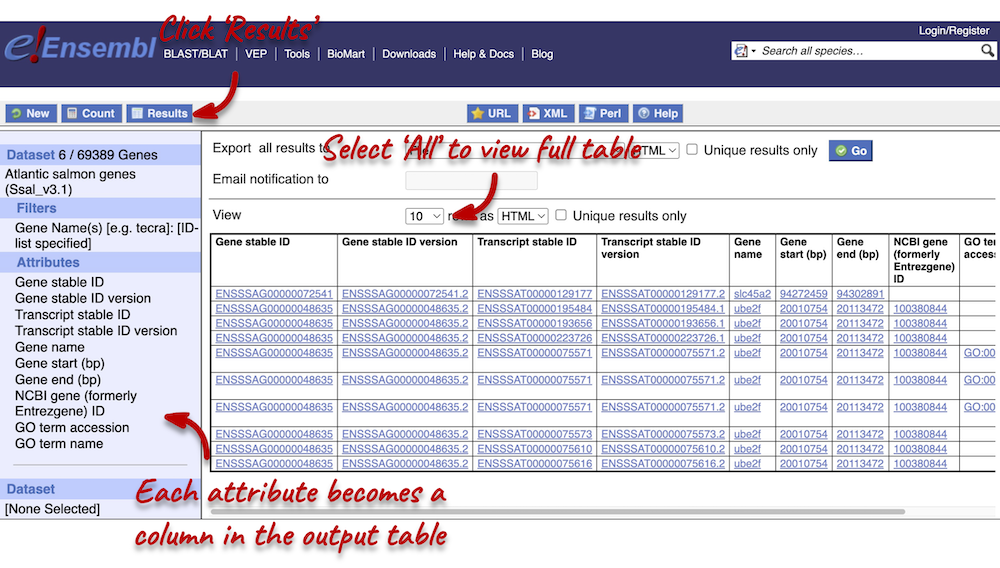
Select All rows from the drop-down menu under View to open all results in a new tab. You will notice that there are multiple results for gene names slc45a2 and ube2f. This is because the names can be ambiguous. If you focus on the Gene stable ID column, you will notice that gene names slc45a2 and ube2f will each have 2 different gene stable IDs assigned to them. Gene stable IDs are unique.
You may see multiple entries for each gene stable ID. This is because a gene can be transcribed into a number of transcripts, so you will have results for each of the transcripts (see the Transcript stable ID column). Finally, you may also notice that there are multiple entries for each transcript. This is because multiple GO terms can be associated to a single transcript (see the GO term accession column).
Step 3.2: Select attributes (sequences)
To complete the third part of the demo, we need to export the cDNA sequences of our three genes. To do this, go back to the Attributes section in the left-hand panel and select Sequences from the attributes categories at the top of the page. Expand the SEQUENCES tab and select cDNA.
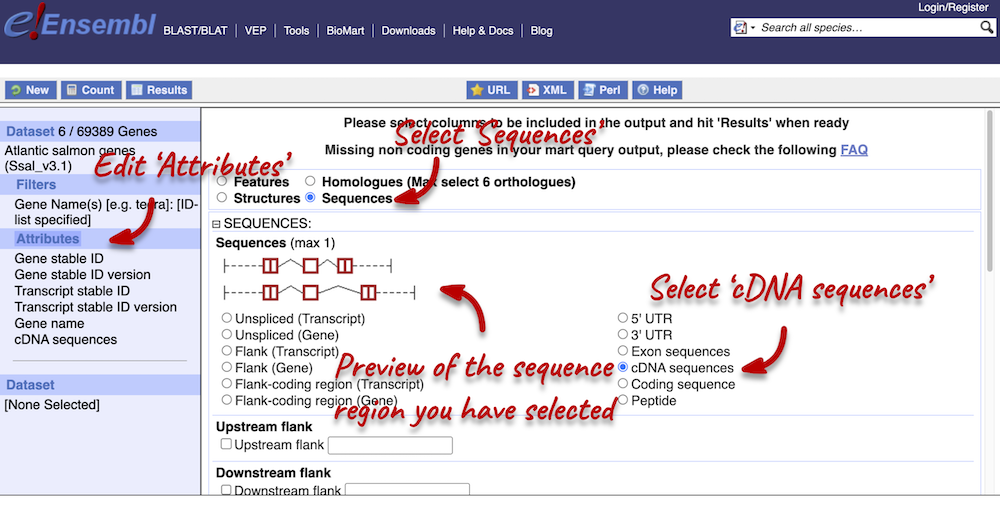
Next, expand the HEADER INFORMATION tab and select Gene name, so that we can match our output to our input again. Click Results in the left-hand panel to see your sequences.
Step 4.2: Get the results (sequences)
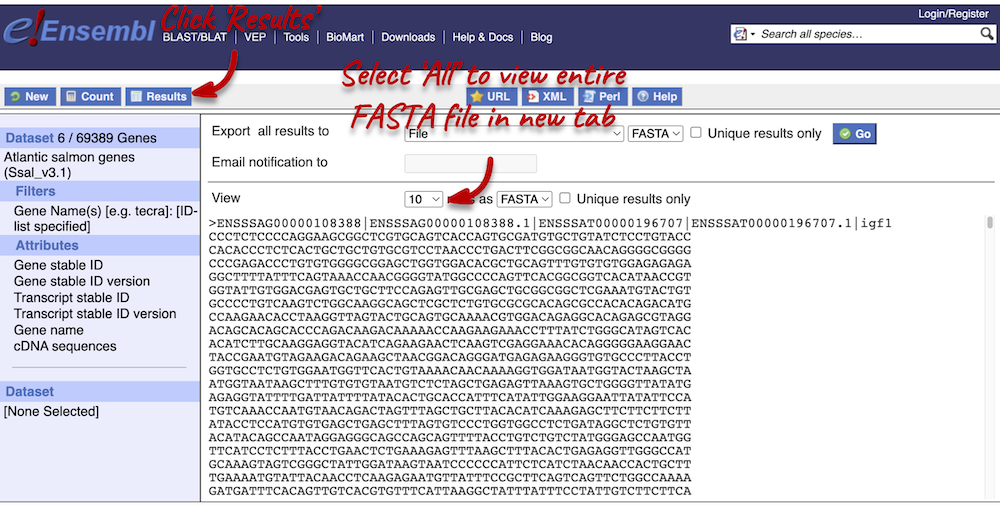
Click on Results again in the top left-hand corner to view your cDNA sequences in FASTA format. You can find your gene and transcript stable IDs, as well as the gene name in your FASTA header, which is followed by the cDNA sequence itself.
For more details on BioMart, have a look at this publication:
Kinsella RJ, Kähäri A, Haider S, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database: the Journal of Biological Databases and Curation. 2011 ;2011:bar030. DOI: 10.1093/database/bar030. PMID: 21785142; PMCID: PMC3170168.






