Whole-genome alignments, Demo
Demo: Whole-genome alignments
Alignments in the Region in Detail view
Pairwise alignments
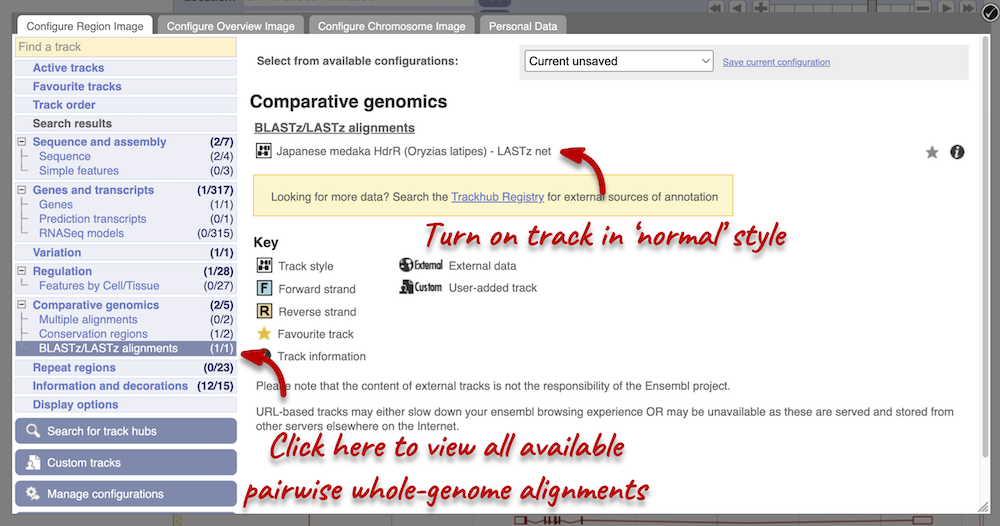
Let’s look at some of the comparative genomics views in the Location tab. Go to the region 21:19789800-19872000 in Salmo salar (Atlantic salmon). This region contains a number of different genes. We want to find out if there are any specific regions that align with the Oryzias latipes (Japanese medaka). To do this, we need to look at a LASTz pairwise alignment between the Atlantic salmon and Japanese medaka species. We can look at individual species comparative genomics tracks in the Region in detail view by clicking on Configure this page. In the Comparative genomics section, go to BLASTz/LASTz alignments and turn on the Japanese medaka track in the normal format. Save and close the menu.
We now have a new track, which shows aligned regions in pink blocks. Gaps in the alignment are displayed as empty blocks. Alignments to a single chromosome are presented in a single row. You will find multiple rows of alignments will be shown if the region aligns to more than one chromosome. Click on an aligned region. From the pop-up menu we can see that region of interest only aligns to chromosome 21 in Japanese medaka, and therefore only one row of alignment is shown in the display
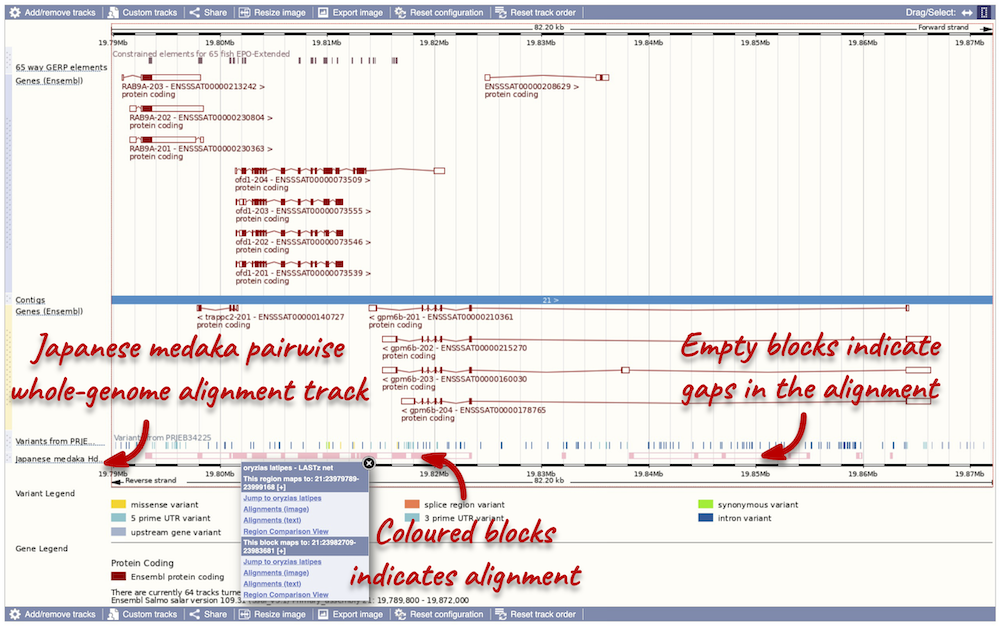
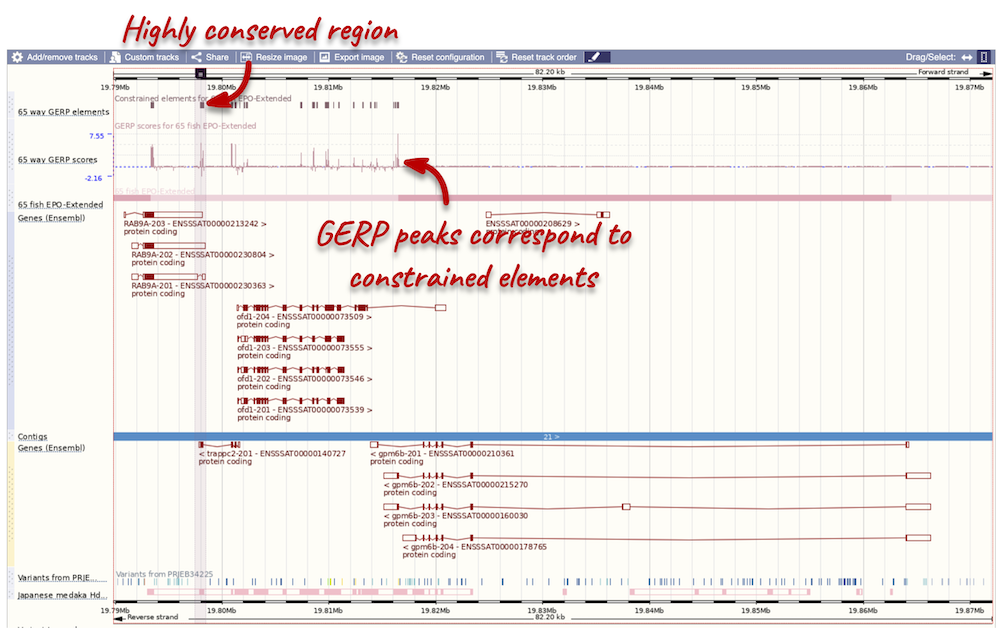
Highly conserved region across the 65 fish EPO-Extendes set align with the
Multiple alignments
Atlantic salmon is part of the 39 fish EPO and 65 fish EPO-Extended comparative genomics analyses. You can view multiple whole-genome alignments on the Region in detail view. Configure this page, go to Comparative genomics: Multiple alignments and turn on the 65 fish EPO-Extended track. Also turn on the Conservation score for 65 fish EPO-Extended and Constrained elements for 65 fish EPO-Extended under Comparative genomics: Conservation regions. You can read more about GERP scores in the Ensembl Comparative Genomics documentation pages. This will show us if any regions are highly conserved within the species set. Save and close the menu.
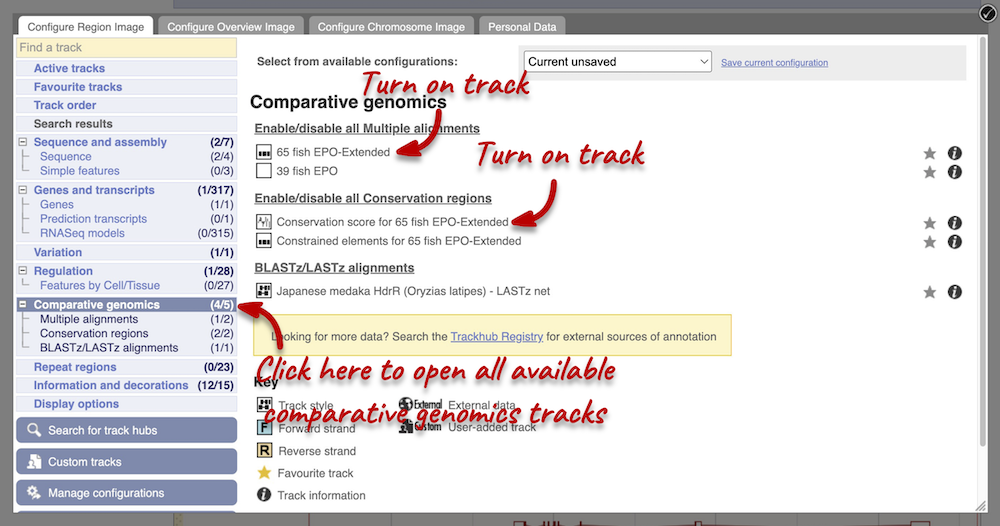
Any conserved regions are shown in the 65 way GERP elemenst track. You can find corresponding conservation scores in the 65 way GERP scores track underneath. These tracks indicate regions of high conservation between species, considered to be “constrained” by evolution.
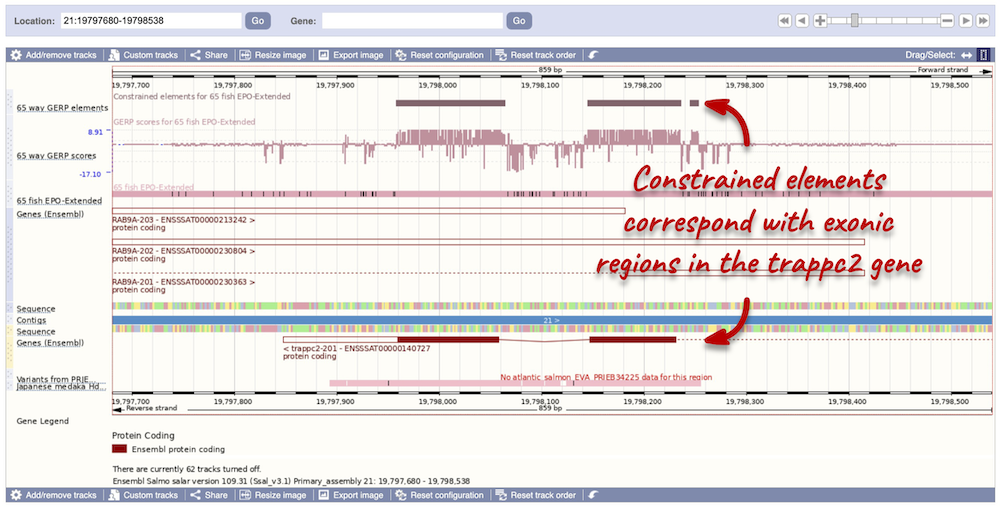
You can zoom into a highly conserved region by dragging your mouse across the region you want to zoom into, and selecting Jump to region in the pop-up menu. Zoomed in, you will be able to see that the constrained elements actually correspond to exonic regions of the trappc2-201 transcript.
Pairwise sequence alignments
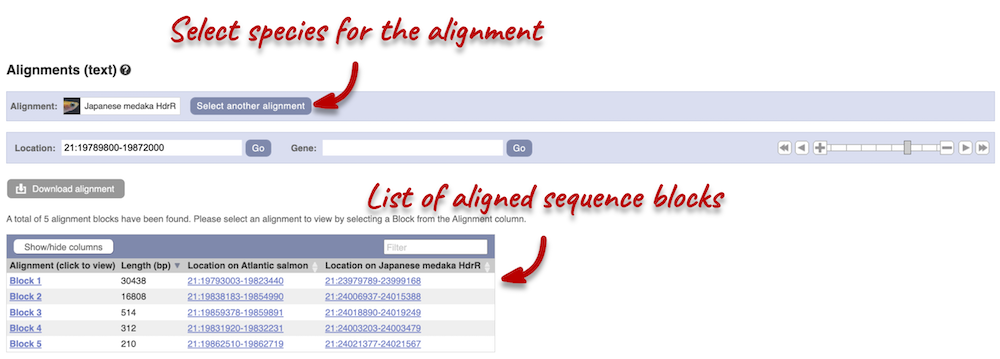
We can look at the nucleotide sequence alignment between Atlantic salmon and Japanese medaka. Click on Comparative Genomics: Alignments (text) in the left-hand menu.
Click on Select an alignment. Under Pairwise, select Japanese medaka and click Apply at the bottom of the menu to save and close. 5 alignment blocks of different lengths are available.
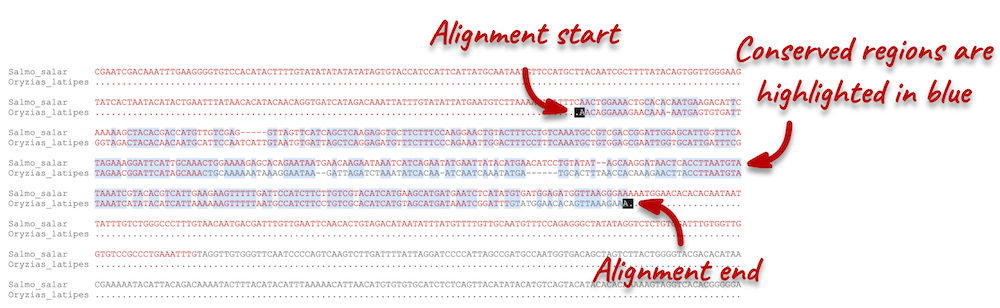
Click on Block 1. You will see a list of aligned regions in Atlantic salmon and Japanese medaka. Scrolling down you will find the nucleotide sequence alignment of the two species. Exons are shown in red.
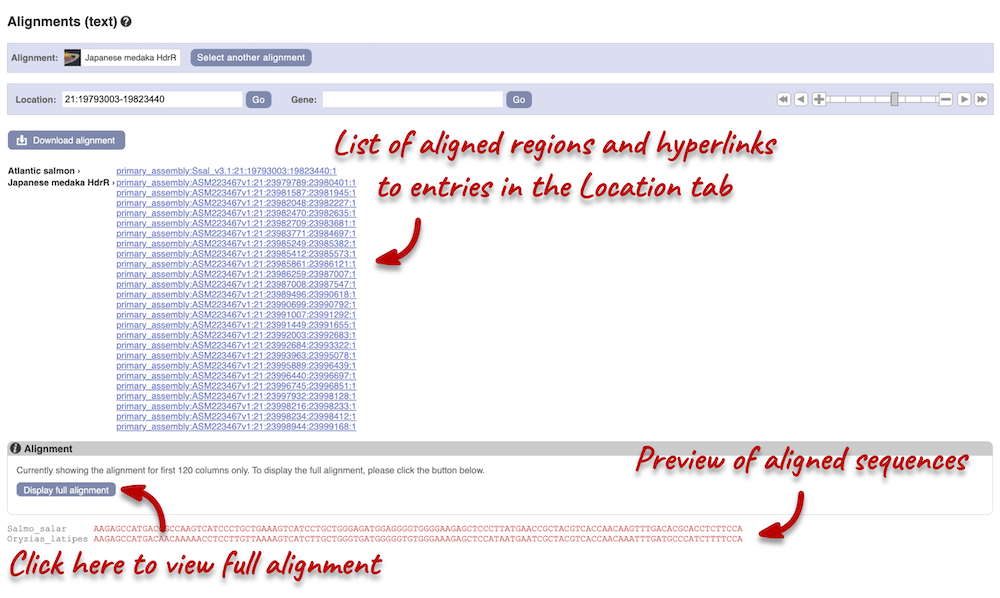
Configure this page on the left. In the pop-up menu, you can turn on the options to view Show conservation regions and Mark alignment start/end. This will add highlights where the sequence matches. Save and close the menu and click on Display full alignment.
Region comparison
To compare the two genomic regions visually, go to Comparative Genomics: Region Comparison in the left-hand panel. To add species to this view, click on the Select species or regions button. Select Japanese medaka again then close the menu. This page, similar to the Region in detail view, shows the chromosome positions (for both species) first. We can see the location of this alignment on chromosome 21 in Japanese medaka. You can scroll down to the Region in detail display. Here, you can view the aligned regions (highlighted in green) between the two species.
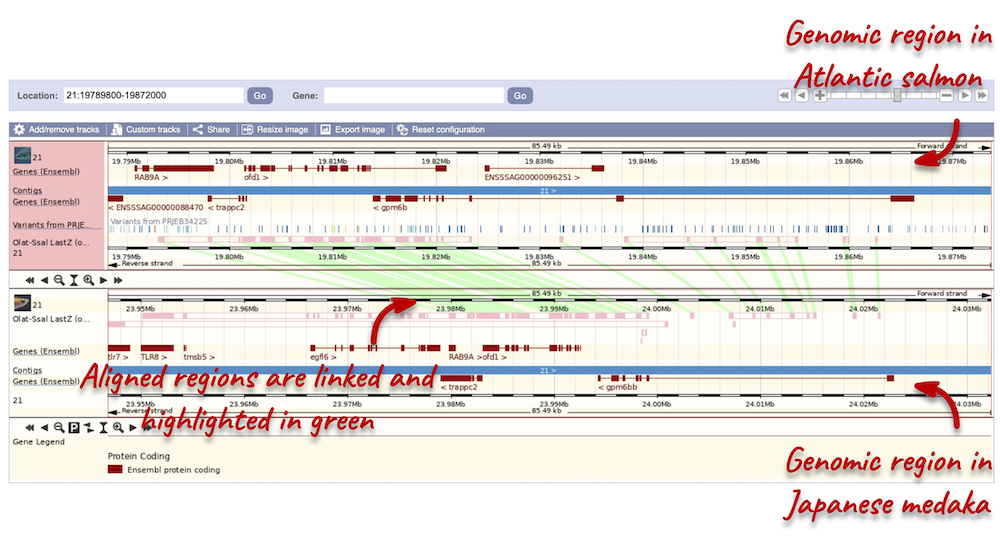
You can add the same tracks as those in the Region in detail view to this region comparison. Click on Configure this page to open the menu and select your tracks of interest.
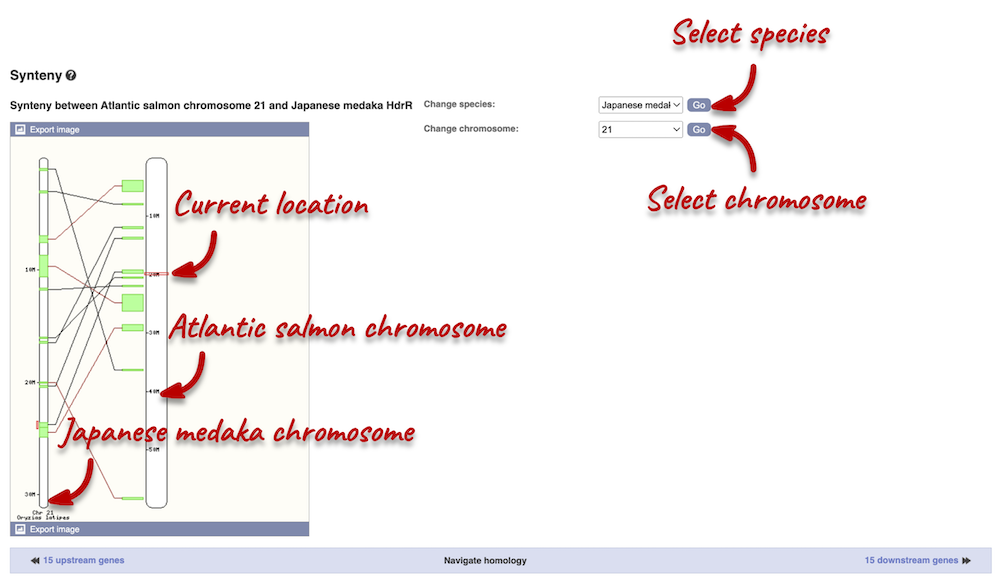
Synteny
We can view large-scale syntenic regions for our chromosome of interest. Click on Comparative Genomics: Synteny in the left-hand panel. Select Japanese medaka from the Species drop-down on the right-hand side. Black linking lines indicate sequences are oriented in the same direction, red linking lines indicate the sequences are inverted.