Exploring a gene in Rice, demo
Now let’s search for a rice gene. Enter OS01G0775500 into the search box on the rice species landing page (below) or in the top right-hand corner of the page.
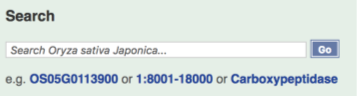
The Search results page displays a single result corresponding to our gene of interest.
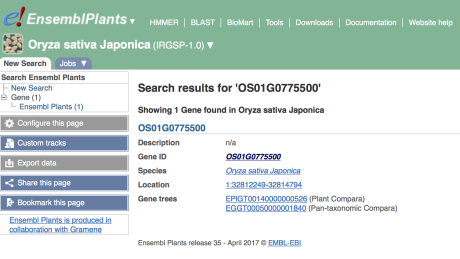
The Gene tab
Click on the link to open the Gene tab for OS01G0775500. On this page you’ll see a table presenting information on the transcript encoded by the OS01G0775500 gene, as well as a graphical model of the gene.
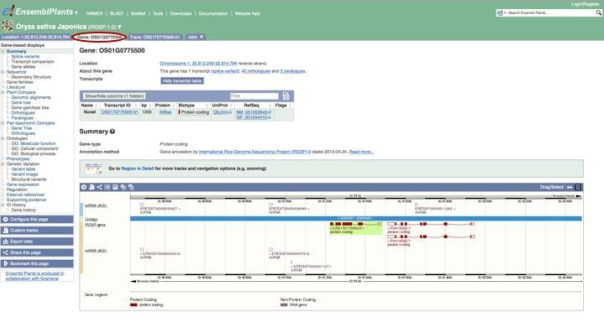
Look through the menu at left to see the different data and annotations available for this gene. You can access sequences; comparative genomics analyses such as alignments, phylogenetic trees and homologue predictions; GO annotations reflecting the function of the protein encoded by the gene; variation data; gene expression data; and links to related data in external repositories.
We’ll start by clicking Sequence to view the FASTA sequence of OS01G0775500.
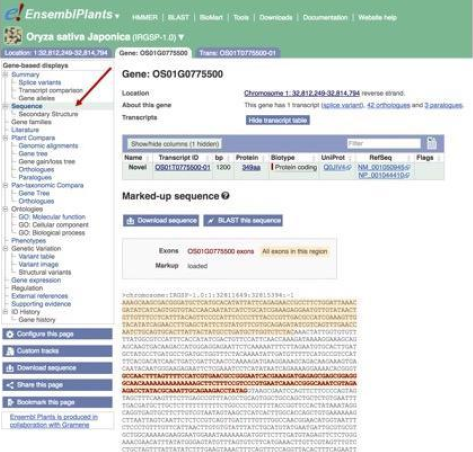
The sequence is marked up such that you can see all exons in the region (orange highlight), as well as exons of OS01G0775500 (bold red font and orange highlighting).
The FASTA header indicates the genome assembly version (IRGSP-1.0), the chromosome (1), the genomic coordinates (32811649:32815394) and the strand (-1, which indicates that the gene is transcribed from the reverse strand).

To download or BLAST the sequence, click the buttons immediately above it.
The Download sequence button is also available below the left-hand menu.

Clicking it brings up a popup showing options to download sequences in FASTA or RTF format (RTF format preserves markup and can be edited in a word processor).
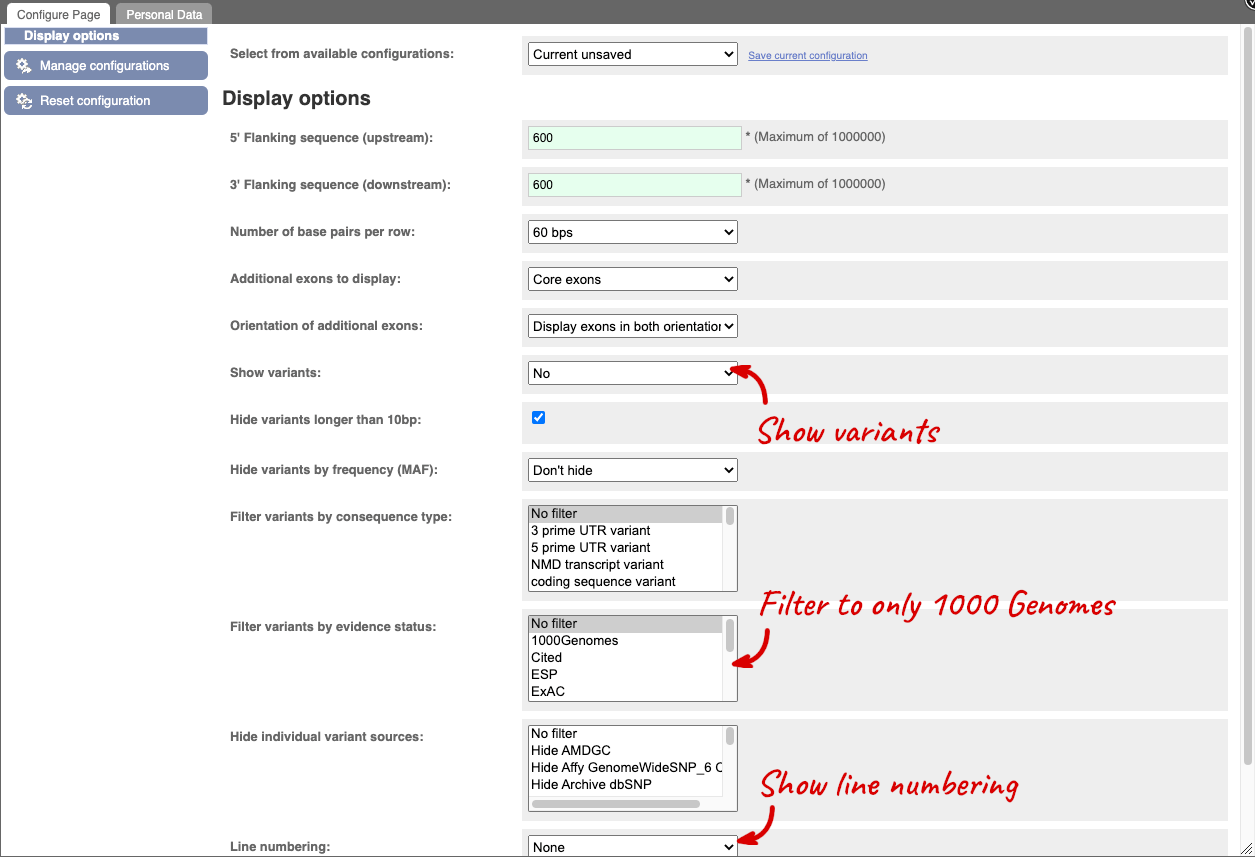
We can change the display of the FASTA sequence by clicking the Configure this page button, at the left:

This loads a popup with a variety of customisation options. For example, you can adjust the amount of flanking sequence displayed, and you can also opt to view variants annotated along the sequence. Click the tickmark in the upper right-hand corner, or anywhere outside the popup, to close the window.
In addition to gene-specific information, we can also access transcript- and genomic-location–specific details.
The Transcript tab
Let’s start by clicking the transcript ID OS01G0775500-01 in the transcript table. This opens the Transcript tab, which presents information specific to the transcript. A full list of available data is accessible from the menu at left.
To view the full sequence of the transcript, click Sequence > Exons:
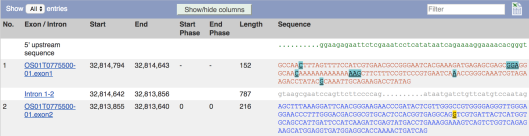
The sequence is colour-coded to indicate whether it is coding or non-coding, and by default, gene variants are identified with coloured highlights. To view spliced sequence, click Sequence > cDNA:
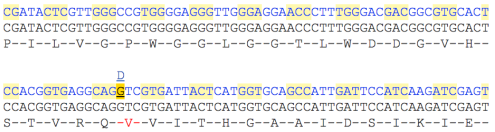
Three tracks, representing the full cDNA sequence (top), the coding sequence (middle), and the translated coding sequence (bottom), are shown. Variant markup is on by default.
Exon and cDNA sequences can be exported by clicking the Download sequence buttons above the sequences.






