Fetching data using the Ensembl REST API, demo
I have the coordinates of a particular protein motif with respect to the protein that it’s in. I would like to find out where this motif lies on the genome.
I’m interested in a coiled-coil domain at position 116-216 in the protein ENSP00000386200.
To do this I want to use the REST API. I’ll start at the REST homepage at http://rest.ensembl.org/.
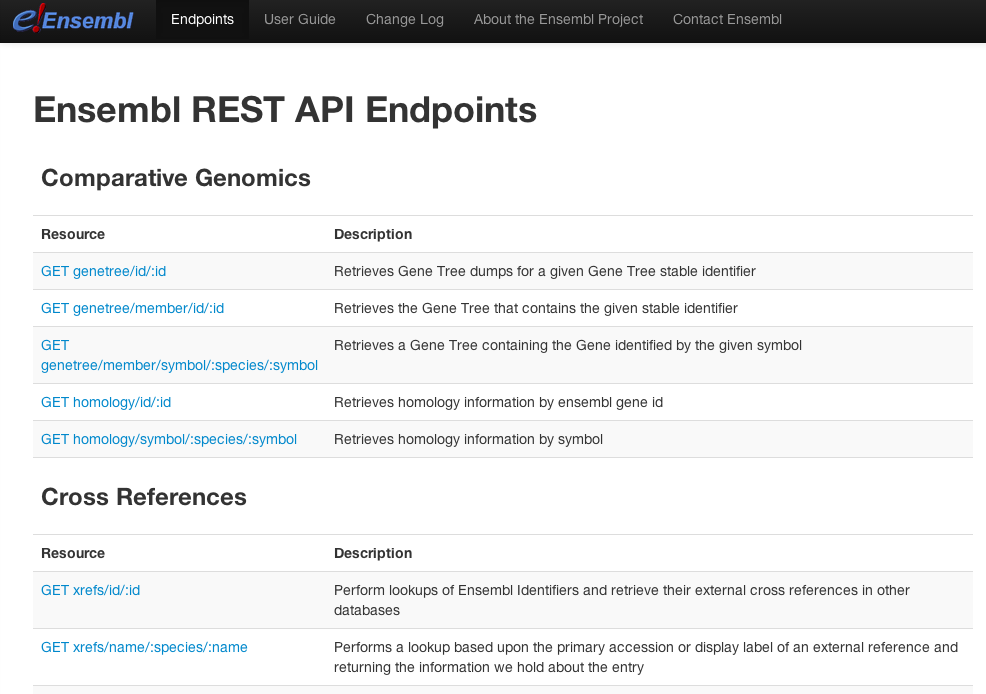
Here you can see a list of all the possible REST endpoints, with names and short descriptions. Scroll down to find the section Mapping. The endpoint GET map/translation/:id/:region does what we want. Click on the link.

If you wish to extract this data using a language such as Perl, Python, Ruby or Java, or to get the data using command line tools such as Curl or Wget, you can click on them to see code examples. We’re just going to do a simple lookup using a URL.
The top of the page shows us that the method is map/translation/:id/:region_. That means that we can get our data using a URL in the format ensembl.rest.org/map/translation/:id/:region.
We also need to set the content type as json, which we do by appending ?content-type=application/json onto the URL.
For our data we can use the URL http://rest.ensembl.org/map/translation/ENSP00000386200/116..216?content-type=application/json. Put this into your internet browser.
This will take you to a text page:
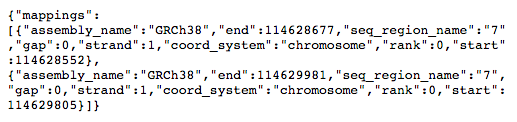
From this we can see that our coiled-coil domain covers two different regions, which will be two different exons of the transcript. They are on chromosome 7 and span 114268607-114268732 and 114269860-114270036.
If we were accessing this data programmatically, the standard json output format would allow us to extract the data.







