Accessing newly annotated genomes in Ensembl Rapid Release, Demo
Newly annotated genomes are all added to Ensembl Rapid Release. These genomes have minimal annotation, with genes mapped onto the genome, InterProScan protein domain analysis and BLAST indexing. There is no BioMart and no comparative genomics analysis. Since gene name and function is usually inferred from orthologues in better annotated species, this is not available for species in Rapid Release.
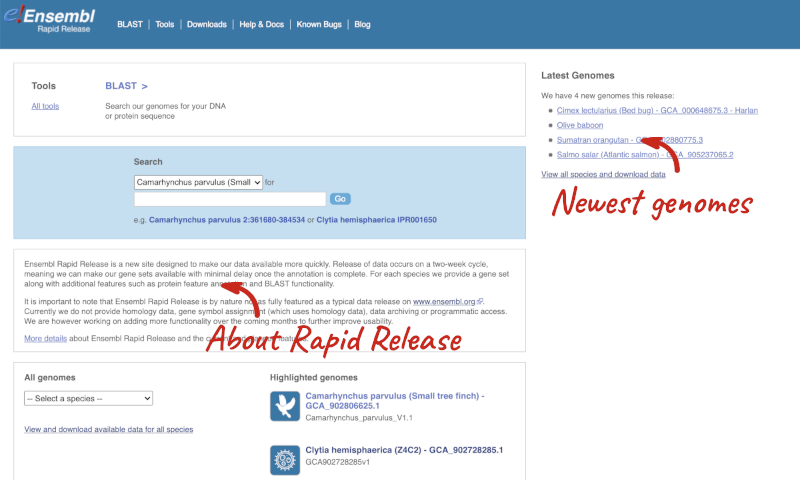
From the Rapid Release homepage, click on View and download available data for all species.
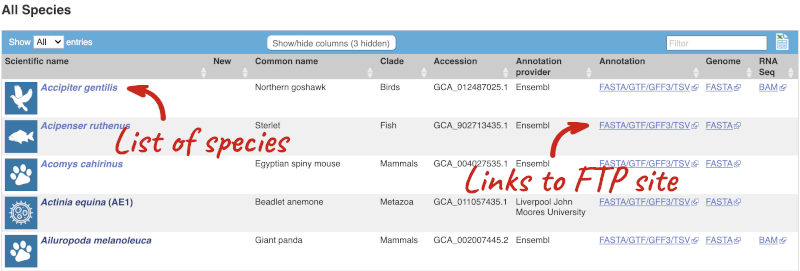
Here we have a list of all the species available in Rapid Release, including latin and common names, genome accession and annotation source. There are also links out to the FTP site where you can download whole genome flatfiles. There are annotation files with the loci and sequences of the genes, the whole genome sequence and BAM files. The BAM files are the RNA-seq data that was used to annotate the genes.
Click on BAM for Ailuropoda melanoleuca (Giant panda). In many internet browsers, this will try to open in an FTP client, so you may need to change the ftp at the beginning of the URL to http to open the site.

Here you can see the BAM files (.bam), BAM index (.bam.bai) and read coverage wiggle files (.bam.bw) for each of the tissues that were sampled for RNA-seq.
Go back to the species list and click on the panda latin name Ailuropoda melanoleuca.
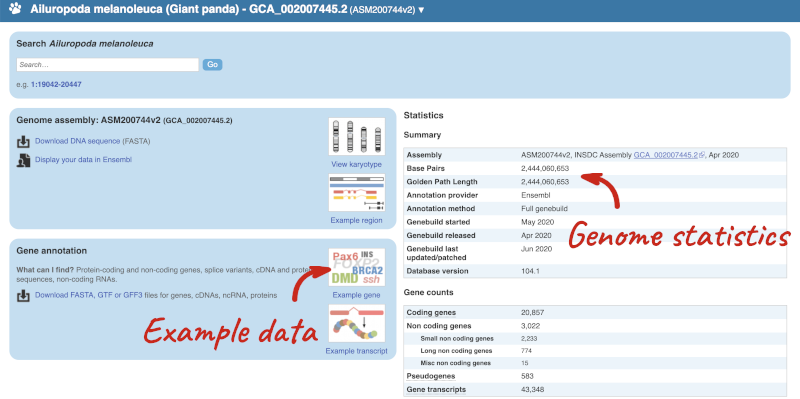
Here you can see links to example data and statistics about the genome. Let’s search for a gene: ENSAMEG00000015675.
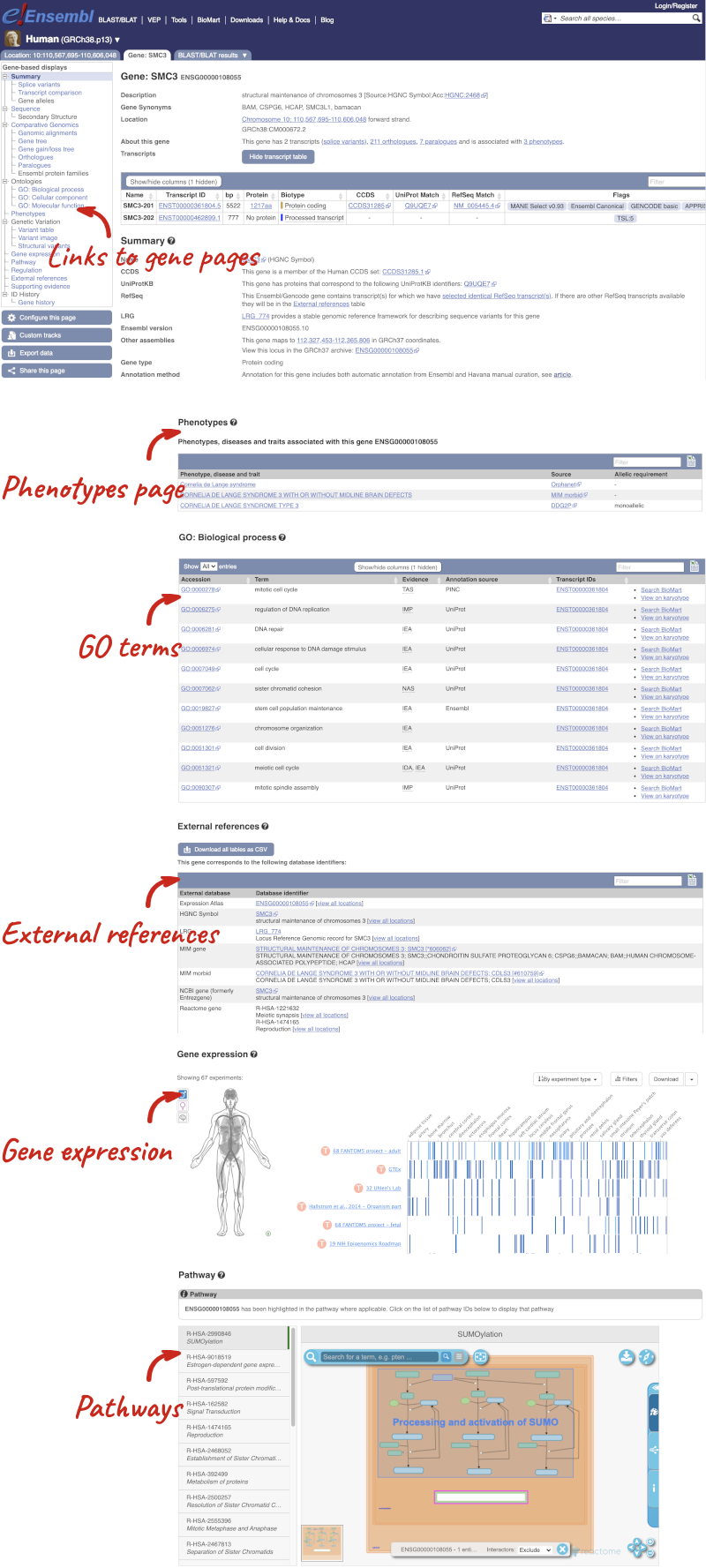
The gene page has a summary of the gene information, including a graphic and table of the transcripts.
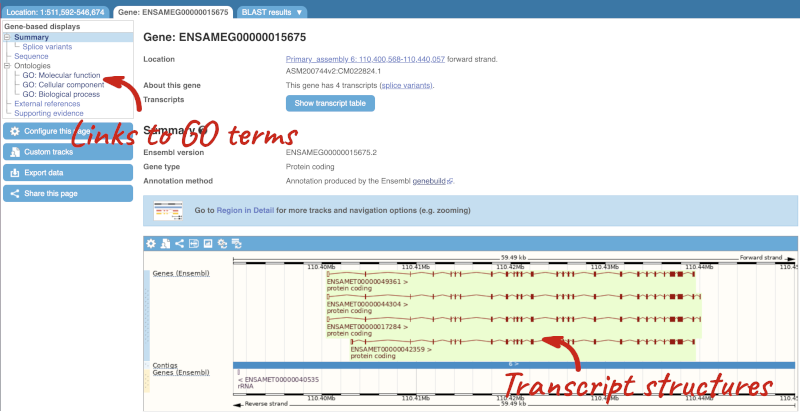
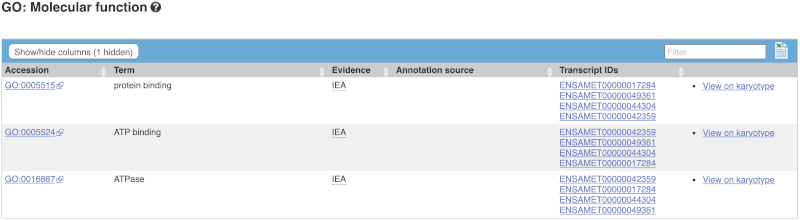
There is a limited amount of annotation attached to the gene. There are GO terms, which were assigned based on protein domains from sequence analysis. Click on any of the GO categories to see the terms.
Let’s explore the Location tab by clicking on Location: 1:511,592-546,674 at the top left. You can view RNA-seq data across this locus by clicking on Configure this page. This menu shows us all the possible tracks you can view. Click on RNA-seq models in the left-hand menu.
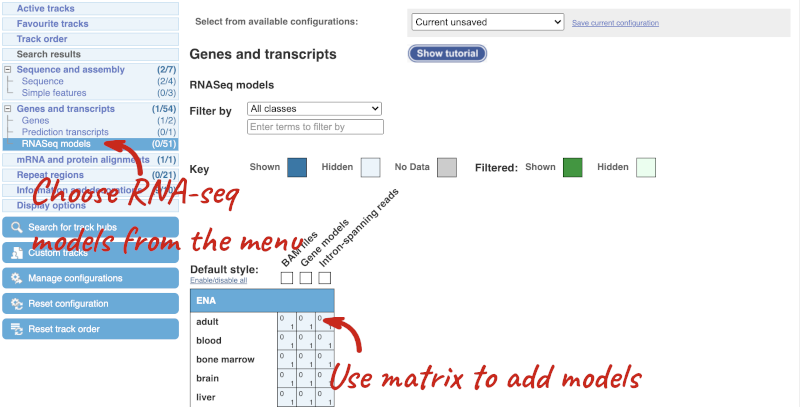
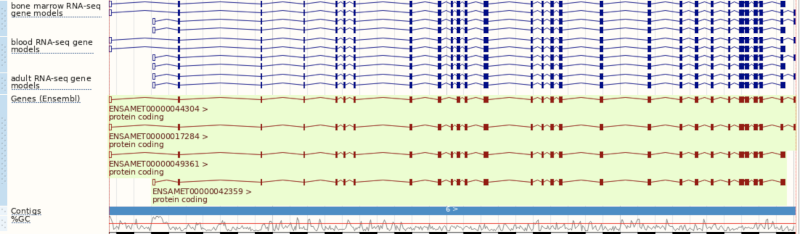
You can use the matrix to add data by cell type. The cell types are listed down the side, with data type along the top. Click on the boxes to add the data individually, or hover over the keys at the top or side to get the option to select all. Hover over Gene models then pick Select all to see all RNA-seq based gene models in different cell types, which were used to annotate genes at this region
One way we can find out more about the gene by BLAST-ing the sequence against a more well annotated gene. We need to export the sequence first. First go the gene tab, then expand Show transcript table. Click on the transcript ENSAMET00000049361.1. This will take you to the transcript tab, where you can click on Protein to get the protein sequence. Copy the sequence and launch www.ensembl.org, then go to BLAST/BLAT.
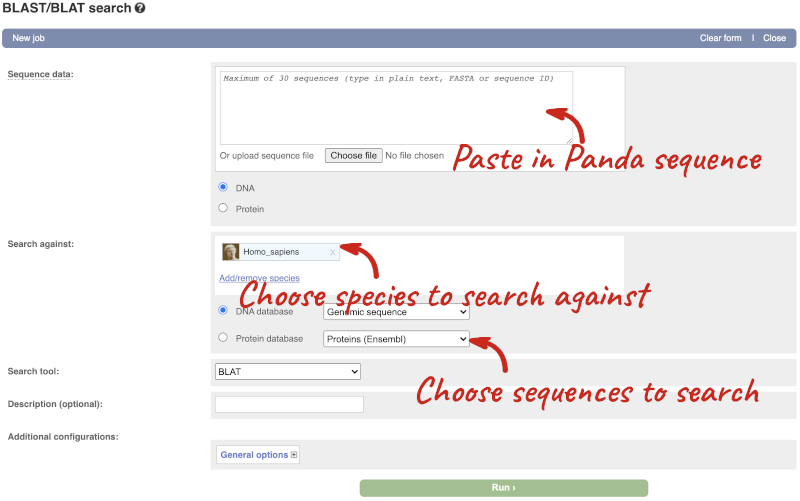
Paste in the Panda sequence, then search against the Protein database and hit Run. Go to the results when the job is done.
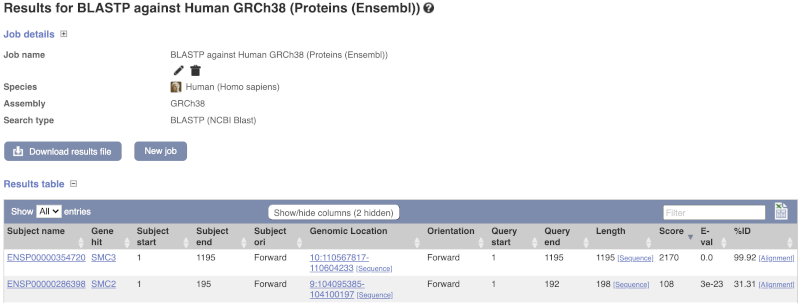
The top hit is to human SMC3, which we can assume is the orthologue of the Panda gene. Click on the gene to go to the gene tab. You can find significant information about the human gene from the gene tab, much of which probably applies to the Panda gene too, for example Phenotypes associated with the human gene, GO terms linked to the human gene, External references to other databases, tissue-based Gene Expression from Expression Atlas and biochemical Pathways the protein is involved in from Reactome.