Custom data, Demo
Demo: Upload small files
We have some Arabidopsis mutants characterised by increased plant branching. They all have large scale deletions on chromosome one:
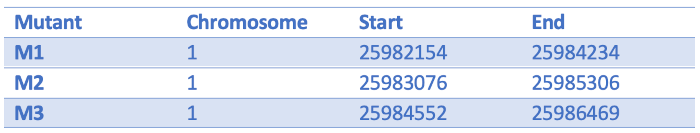
We can turn them into a BED file and view them in the genome browser:
chr1 25982154 25984234 M1
chr1 25983076 25985306 M2
chr1 25984552 25986469 M3
You can add data from a Region in Detail page by clicking on the Custom tracks button at the left. Alternatively, go to a species homepage and click on Display your data in Ensembl Plants.

A menu will appear:
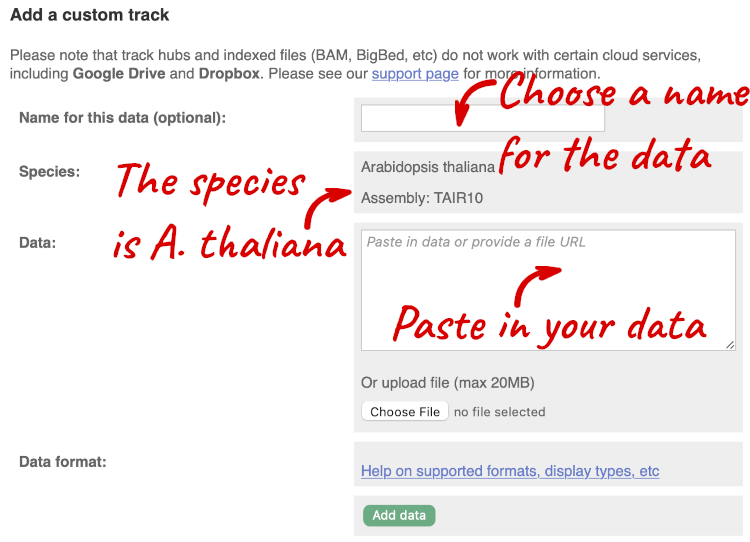
The interface detects file types if you upload or attach a file. When you paste in your data, it can’t do this so we have to tell it what our file type is. It will give you an option where you can select BED.
Click Add data.
You should get to a dialogue box telling you your upload has been successful.
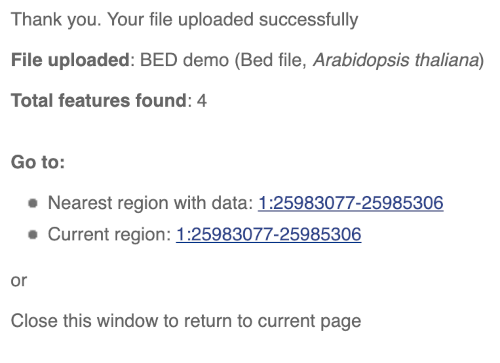
Click on the genomic coordinates link to go to the nearest region with data.
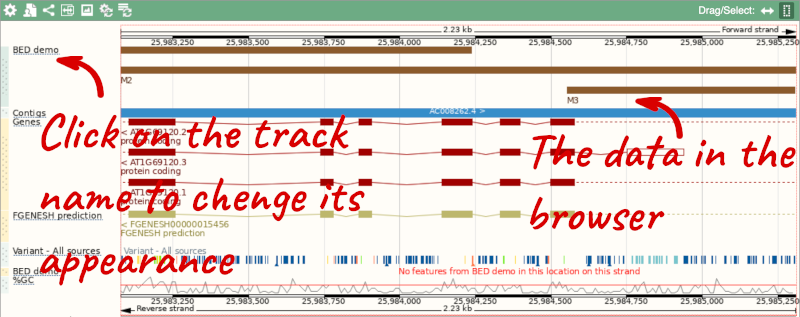
To have a look at the file, click on Custom tracks.
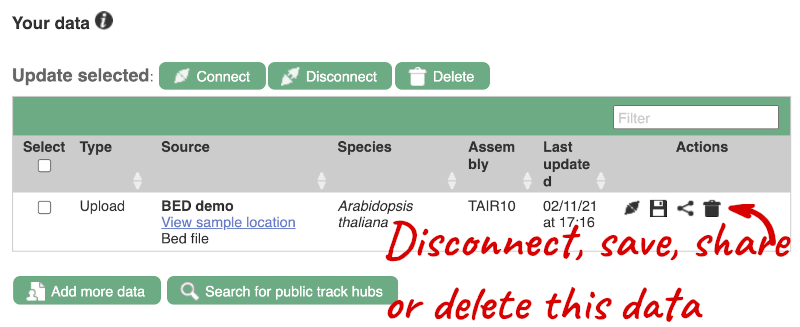
If you’ve got an Ensembl account, you can save this data to your account. Accounts are free to set up and allow you to save configurations and data, and share with groups. You can also permanently delete or temporarily disconnect data from here.
Demo: Attach URLs of large files
Larger files, such as BAM files generated by NGS, need to be attached by URL. You can find seedling RNAseq reads from the 19 genomes project aligned to the Arabidopsis thaliana assembly here: http://mtweb.cs.ucl.ac.uk/mus/www/19genomes/RNA.seedlings.BAM/v9/Col_0.R1.9.bam
Let’s take a look at the folder.
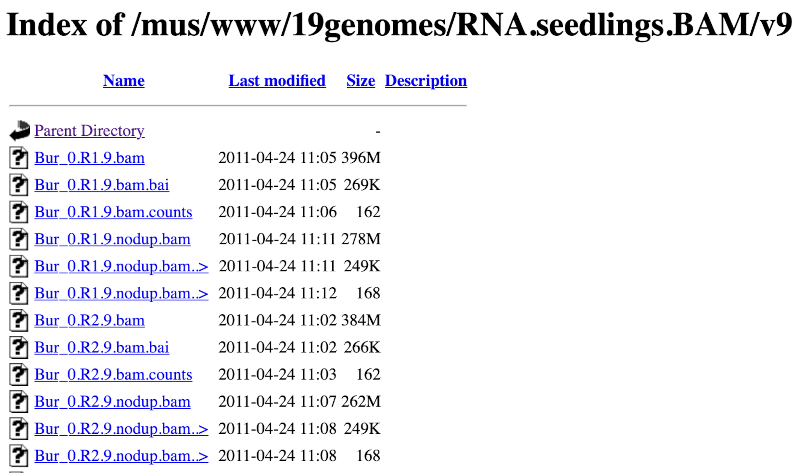
Here you can see a number of BAM files (.bam) with corresponding index files (.bam.bai). We’re interested in the files Col_0.R1.9.bam and Col_0.R1.9.bam.bai. These files are the BAM file and the index file respectively. When attaching a BAM file to Ensembl, there must be an index file in the same folder.
To attach the file, click on Custom tracks, then click on Add more data to add a new track.
We get to the same dialogue box as before. This time we’ll name our data RNAseq reads.
Paste in the URL of the BAM file itself (http://mtweb.cs.ucl.ac.uk/mus/www/19genomes/RNA.seedlings.BAM/v9/Col_0.R1.9.bam).
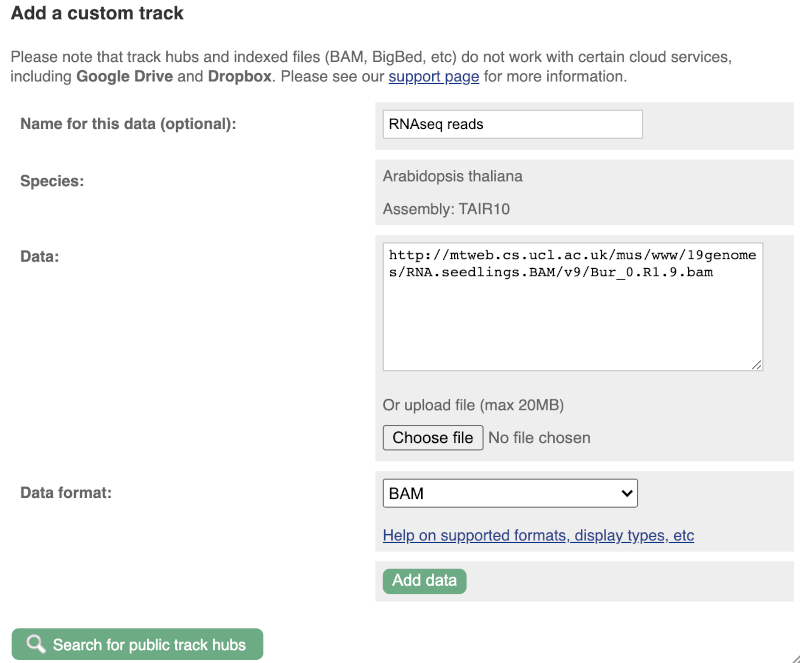
Since this is a file, the interface is able to detect the “.BAM” file extension, so automatically labels the format as BAM. Click on Add data. You should get to a dialogue box telling you your data has been attached successfully. Close the menu to go back to your region of interest.
Let’s go to the region of the AT1G51745 gene. Search for the gene using the Gene text box and click Go.
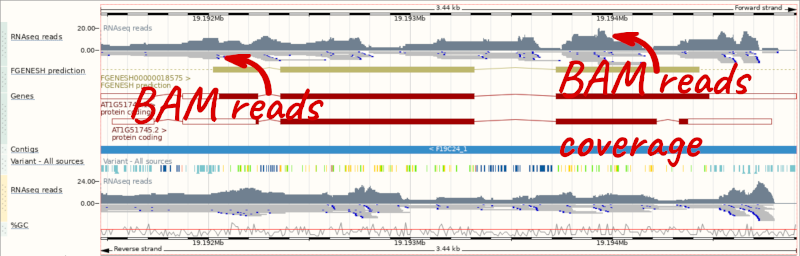
We can zoom in to see the sequence itself. Drag out boxes in the view to zoom in, until you see a view like this. Alternatively, type 1:19194595-19194630 in the Location box and clicking Go to jump to a smaller region.
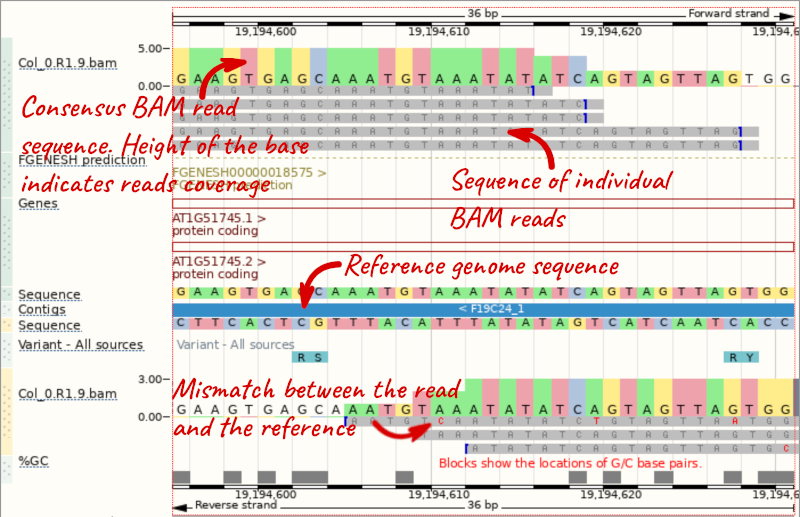
Any mismatches between the reads and the reference genome assembly are shown in red.
Demo: Track hub registry
Track Hub Registry provides publicly available data organised in track hubs. Ensembl established a pipeline for generating track hubs for all public RNA-Seq studies in the INSDC archives. This pipeline discovers and aligns reads from RNA-Seq studies across all plant species in Ensembl Plants, which means that you can search the Track Hub Registry for available RNA-Seq data and display them in the genome browser.
You can search for track hubs to add in different ways:
- Search for track hubs in the Track Hub Registry and choose to add them to your genome browser of choice.
- Search the track hub registry using the Track Hub Registry interface in Ensembl Plants (there is a link from the homepage).
We will now add the track hub containing data on epigenetic regulation of transcription initiation in Arabidopsis (DRP006159).
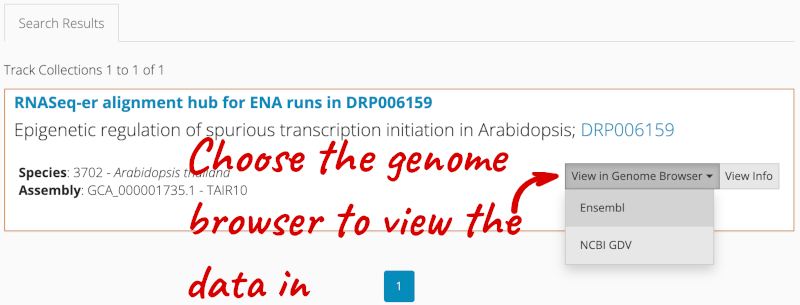
You can add track hubs to view in Ensembl directly via the Track Hub Registry. Go to the Track Hub Registry homepage and search for DRP006159.
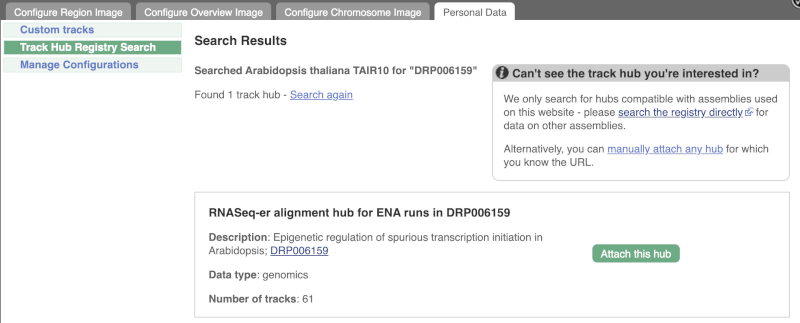
There is one RNA-seq alignment hub returned, which you can view in the genome browser.
Alternatively, you can add track hubs by searching the Track Hub Registry through Ensembl. Click the Custom tracks -> Track Hub Registry Search in any region view within Ensembl.
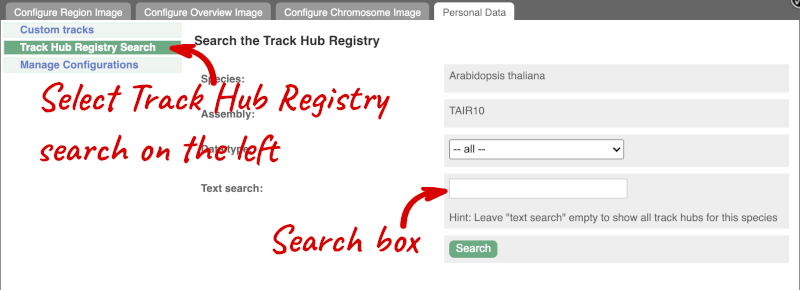
You can only find track hubs for the selected species and assembly denoted in the search box.
Search for DRP006159.
Click Attach this hub in the search results page.
Track Hubs often contain vast amounts of data, which can slow Ensembl down, so only add them if you need them, and trash them when you are finished with them.
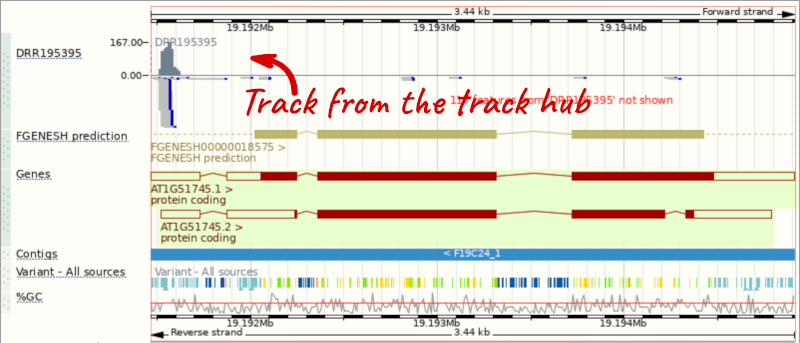
Go to Configure this Page to see that a new category has been added to your menu. Add track for the DRR195395 run to the Region in Detail view by ticking the box.
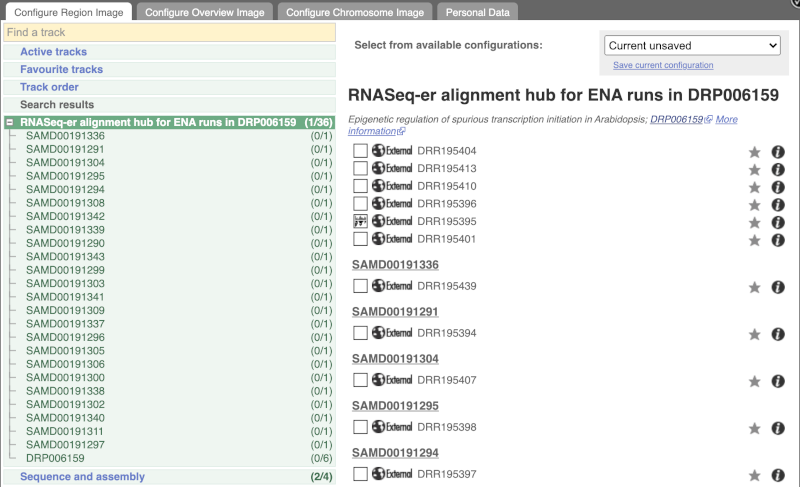
This data represents genome-wide map of transcription start sites (TSSs) in A. thaliana mutants generated using CAGE-seq. Can you see the high reads coverage corresponding to the TSS of our AT1G51745 gene?