Annotating genetic variants with the VEP, Demo
We have identified seven variants in pig:
rs319195925, rs80805426, rs81267388, rs80854621, rs711163915, rs321793337, rs792403417
We will use the Ensembl VEP to determine:
- If the variants have been annotated in Ensembl already
- If genes are affected by the variants
Go to the front page of Ensembl and click on Variant Effect Predictor in the Tools section or click on VEP in the top header.

This page contains information about the VEP, including a link for downloading the script version of the tool. Click on the Launch VEP button to open the input form.
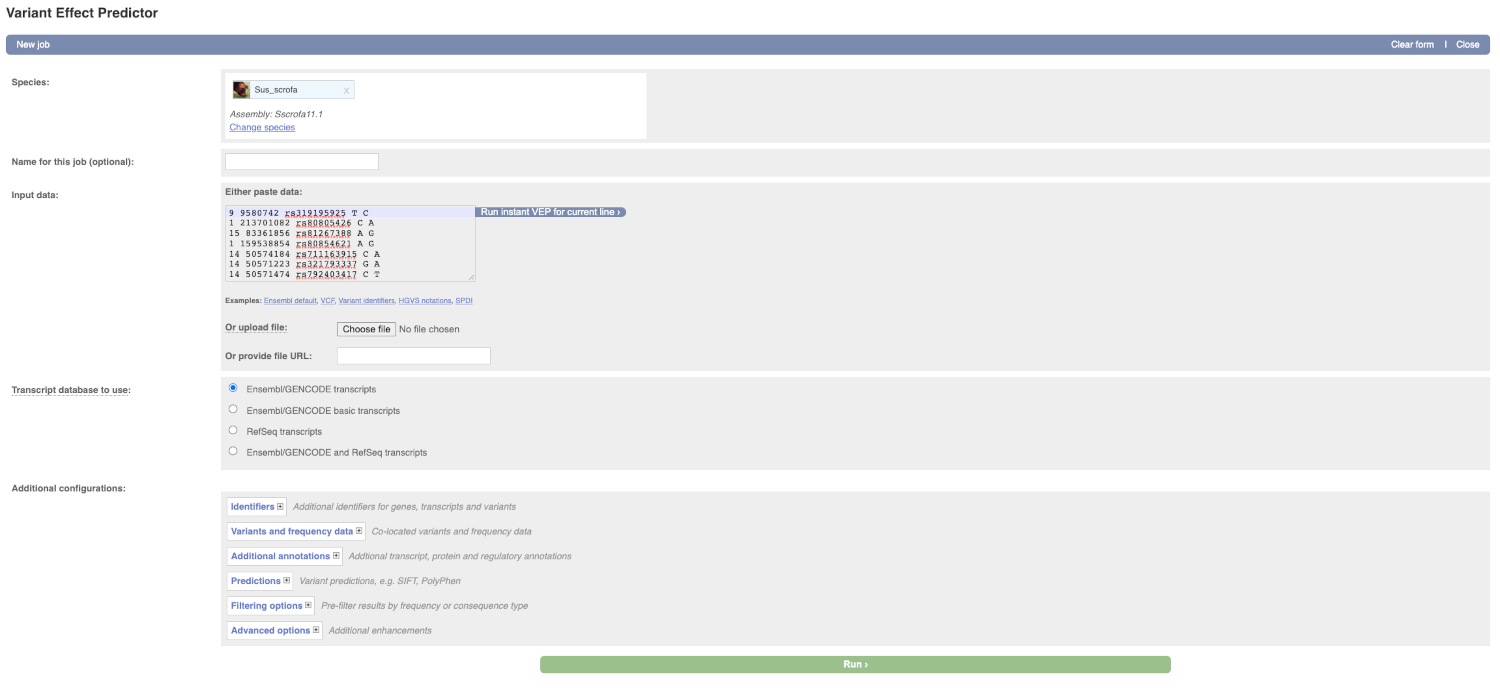
Lets input the variants data in VCF format:
Chromosome Position Name Reference Alternative
Put the following into the Input data box:
9 9580742 rs319195925 T C
1 213701082 rs80805426 C A
15 83361856 rs81267388 A G
1 159538854 rs80854621 A G
14 50574184 rs711163915 C A
14 50571223 rs321793337 G A
14 50571474 rs792403417 C T
The VEP will detect automatically that the data is in VCF format.
There are further options that you can choose for your output. These are categorised as Identifiers, Variants and frequency data, Additional annotation, Predictions, Filtering options and Advanced options. Let’s open all menus and take a look.
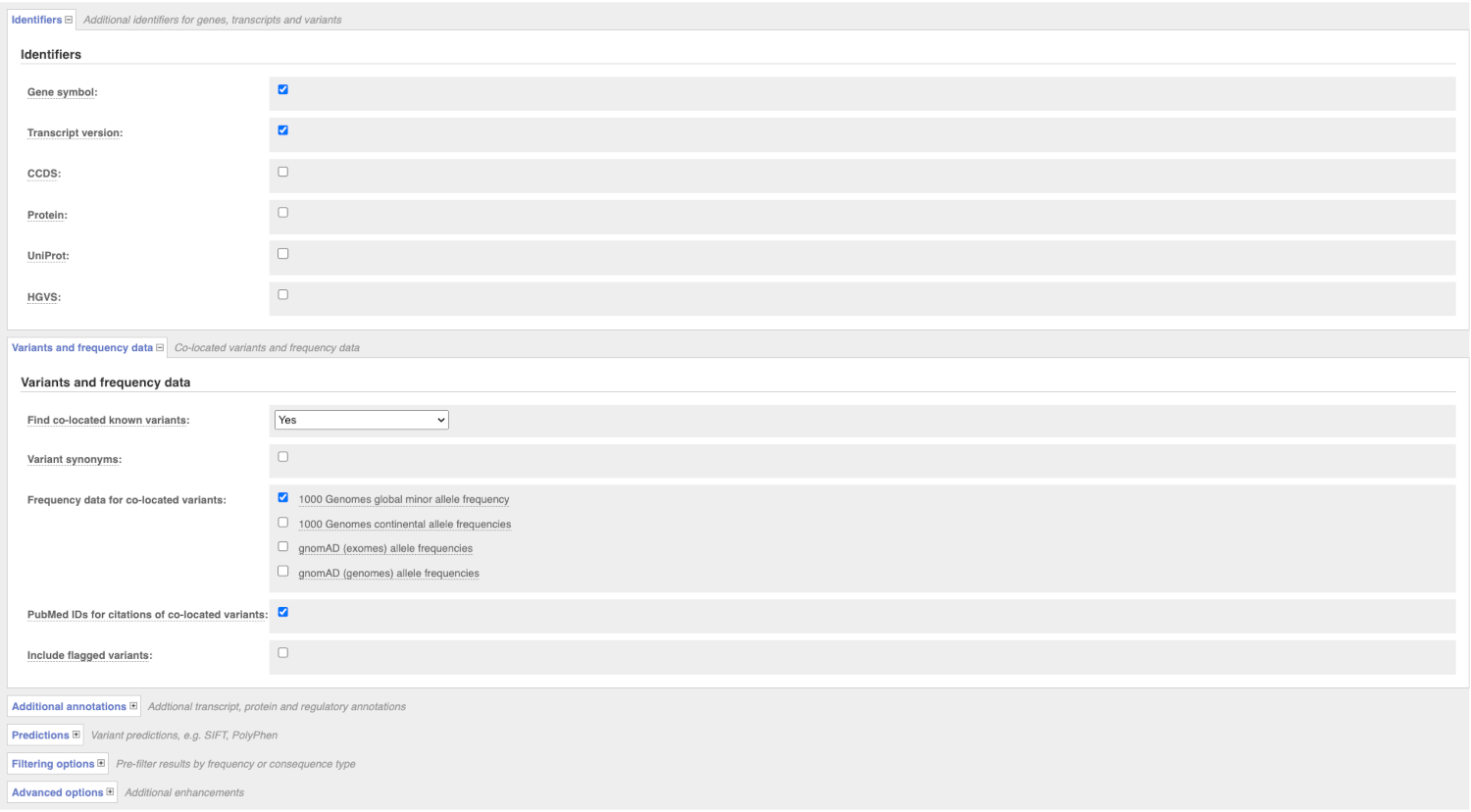
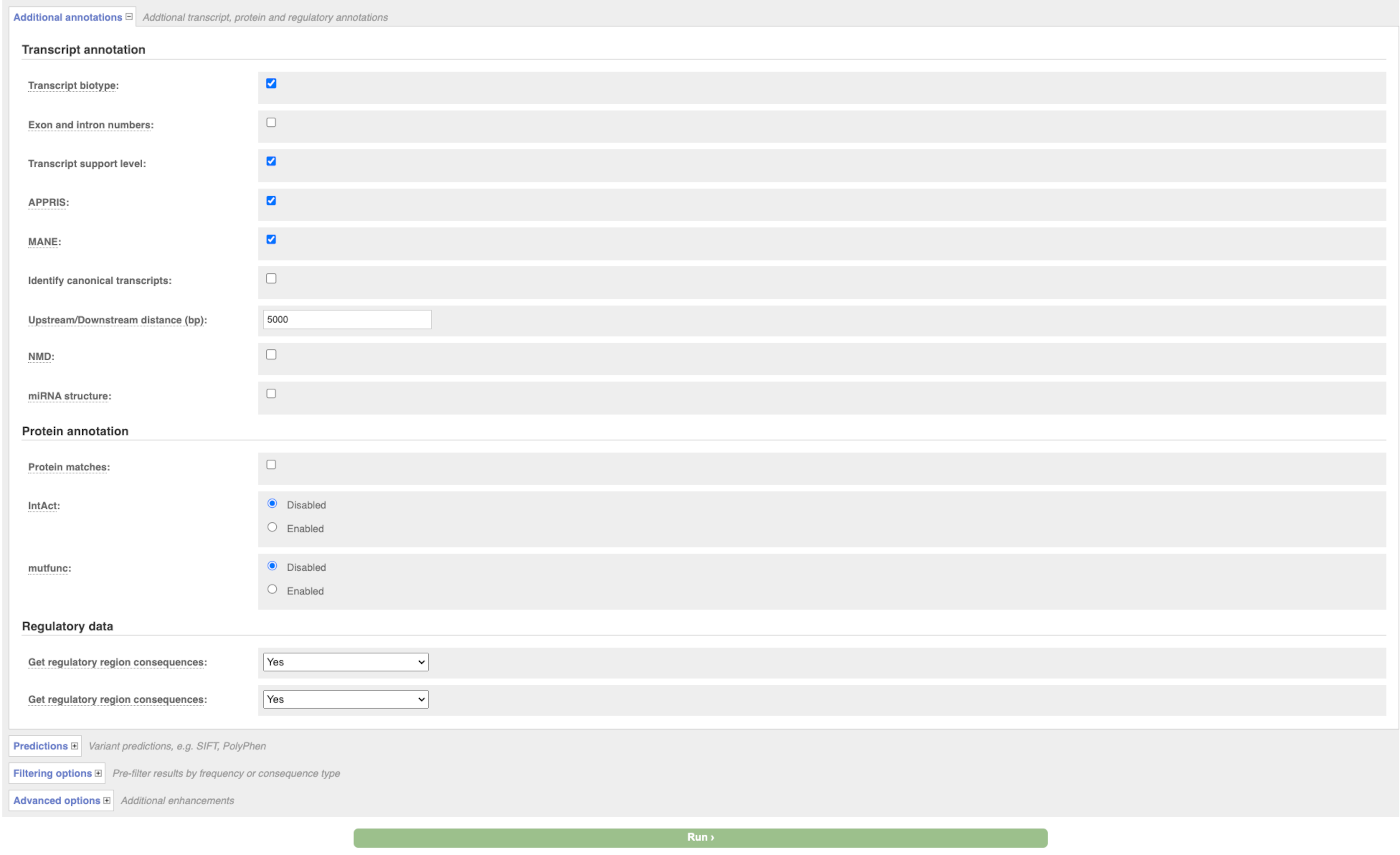
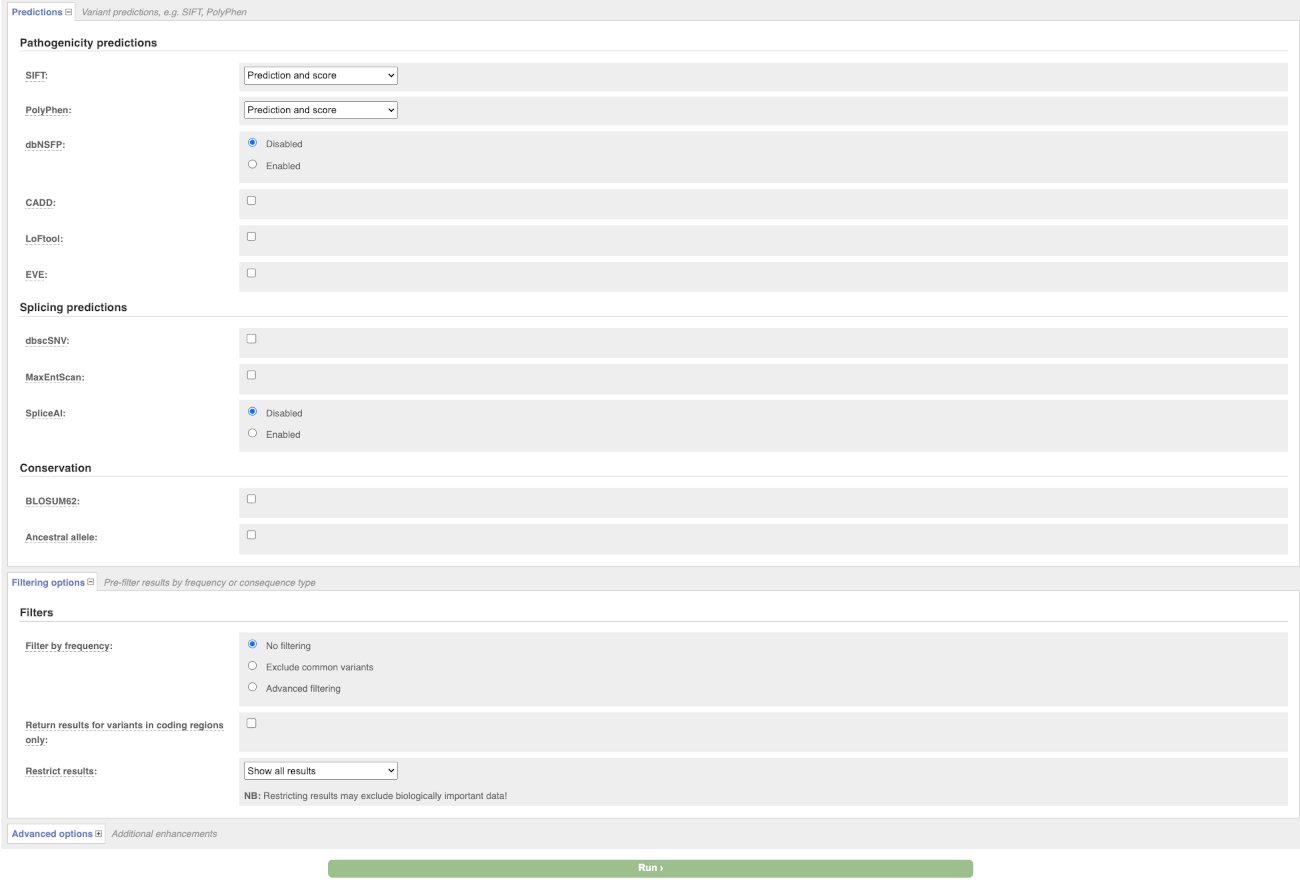
Hover over the options to see definitions.
When you have selected everything you need, scroll right to the bottom and click Run.
The display will show you the status of your job. It will say Queued, then automatically switch to Done when the job is done, you do not need to refresh the page. You can save, edit, share or delete your job at this time. If you have submitted multiple jobs, they will all appear here.

Click on View Results once your job is done.
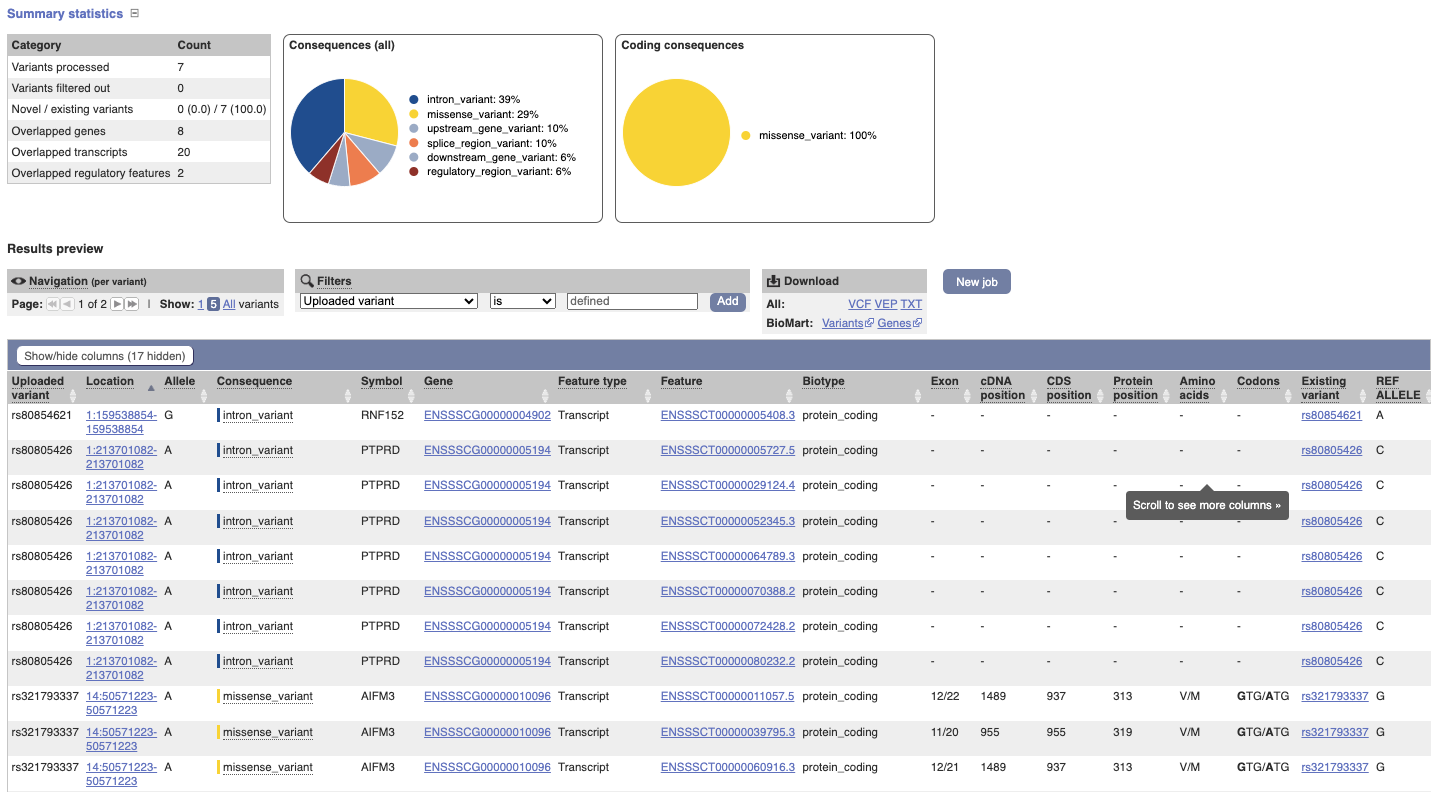
In your results you will see a graphical and table summary of the data as well as a table with the detailed results.