Exporting wheat gene data with BioMart, demo
Demo: BioMart
Follow these instructions to guide you through BioMart to answer the following queries:
- How many genes are found in Fusarium solani that do not have an orthologue in Fusarium oxysporum?
- How many of these are associated with a pathogenic phenotype of “reduced virulence”?
- Export the gene name, locations and GO terms associated with these genes
- Export their cDNA sequences
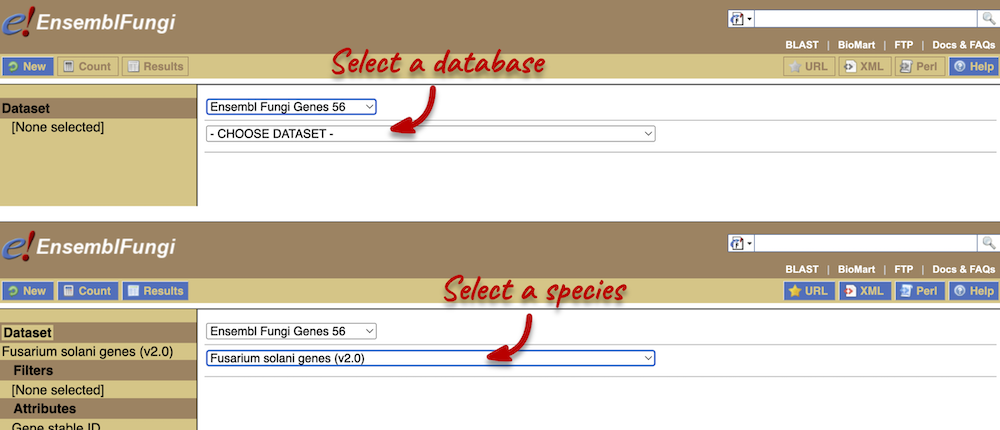
Step 1: Choose database and dataset
Click on BioMart located on the navigation bar at the top of any fungi.ensembl.org page.
Step 2: Choose appropriate filters
We want to find all genes in F. solani that do not have an orthologue with F. oxysporum. We need to filter the dataset to look only at these genes.
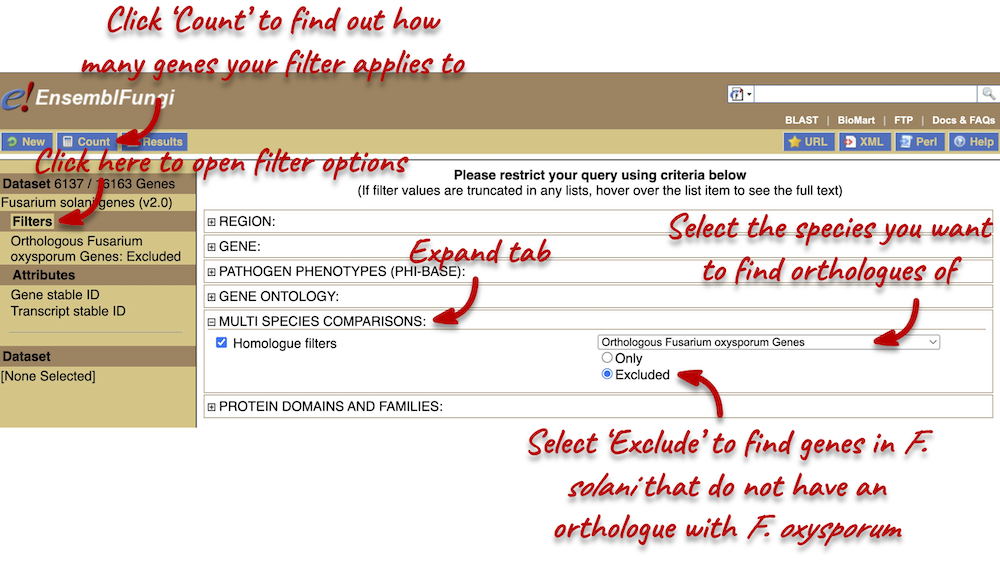
Using the count function we can see that there are 6,137 F. solani genes (out of a total of 16,163) that do not have an orthologue in F. oxysporum. We also want to find out how many of these are associated with the PHI-base Pathogen phenotype ‘reduced virulence’.
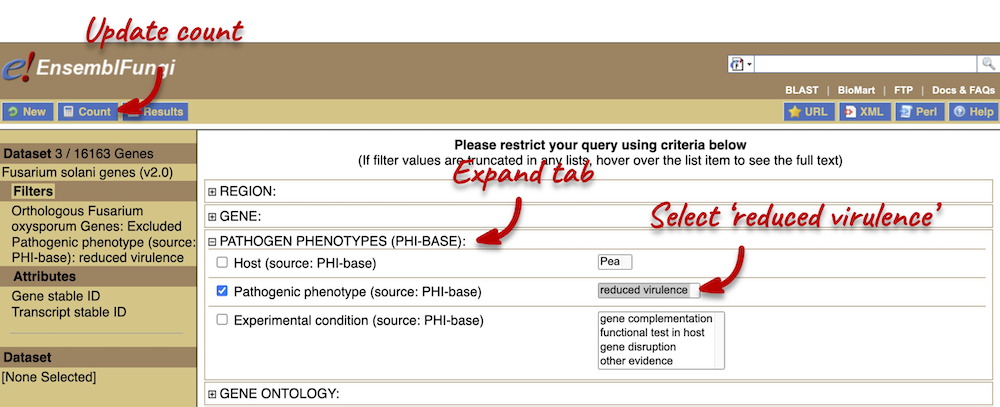
We can now see from the count information that we have 3 genes associated with this phenotype that do not have any orthologues with F. oxysporum.
Step 3.1: Select attributes (features)
Attributes are defined by what we would like to learn about the data. We want to find out more information about the genes:
- Gene name
- Locations
- Associated GO terms
- cDNA sequences
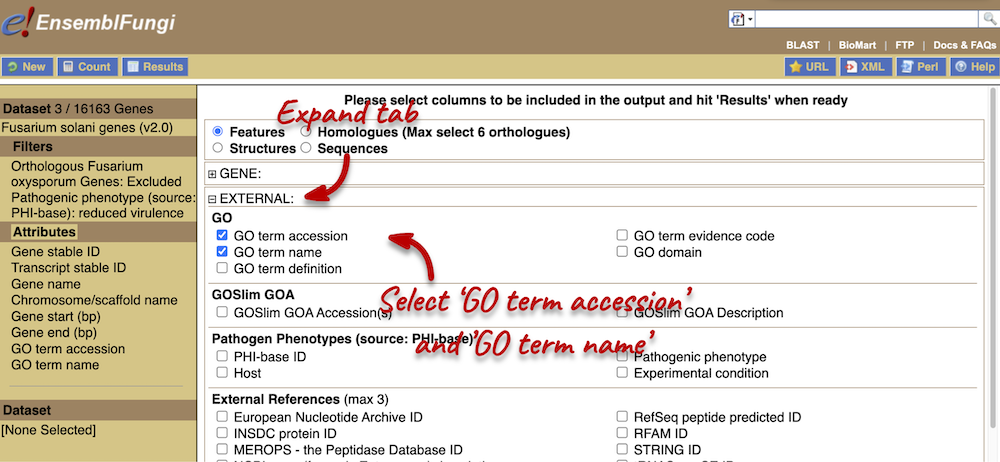
You can only select one attribute category at a time. We can answer points 1-3 in a single query, but we will need to do a second query to answer point 4. Make sure that Features category is selected at the top of the page. Expand the GENE tab and select the following features:
- Gene Name
- Chromosome/scaffold name
- Gene start (bp)
- Gene end (BP)
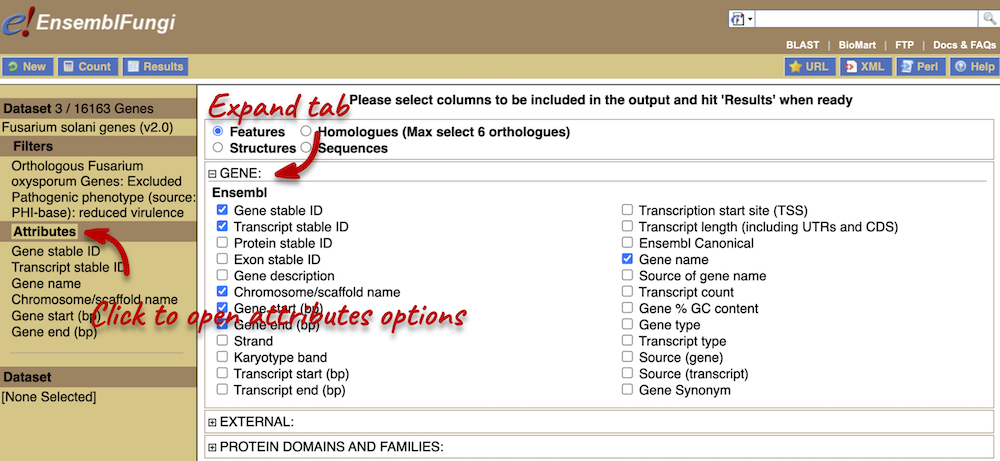
Next, expand the EXTERNAL tab. This section contains lots of identifiers from databases outside of Ensembl. Select the following features:
- GO term accession
- GO term name
Step 4: Get the results
You can download the data if you desire. The table presented shows a sub-sample of 10 results to enable you to check you have the correct attributes.
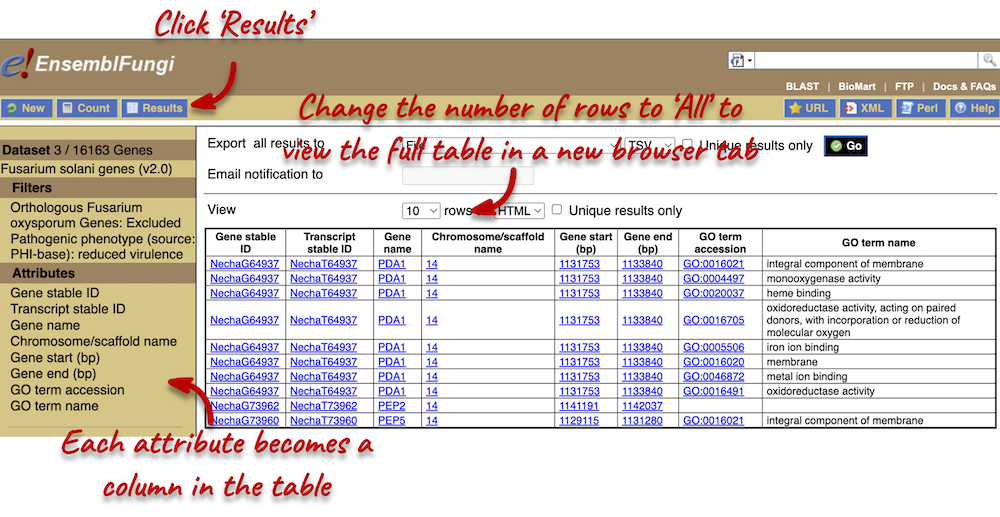
We can see that all of these genes are located in the same region. Perhaps this has something to do with why they are not found in F. oxysporum…? You can click on the location links and explore the synteny between the two species.
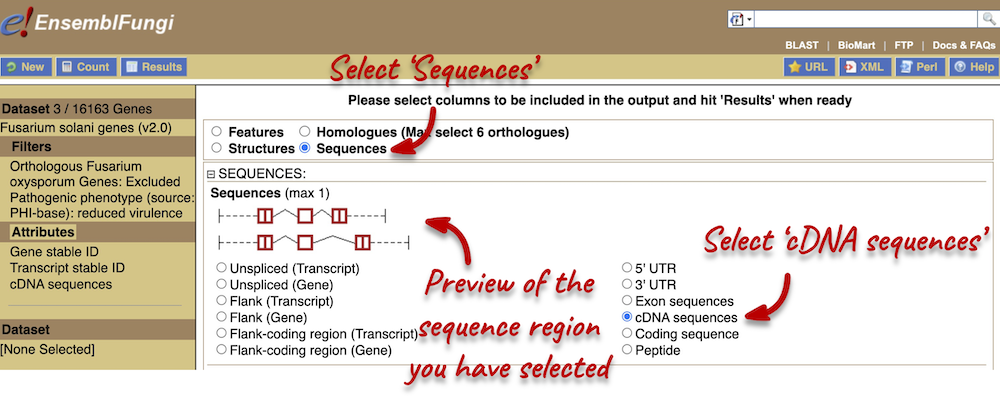
Step 3.2: Select attributes (sequences)
To complete the fourth part of the demo, we need to export the cDNA sequences of our three genes. To do this, go back to the Attributes section in the left-hand panel and select Sequences from the attributes categories at the top of the page.
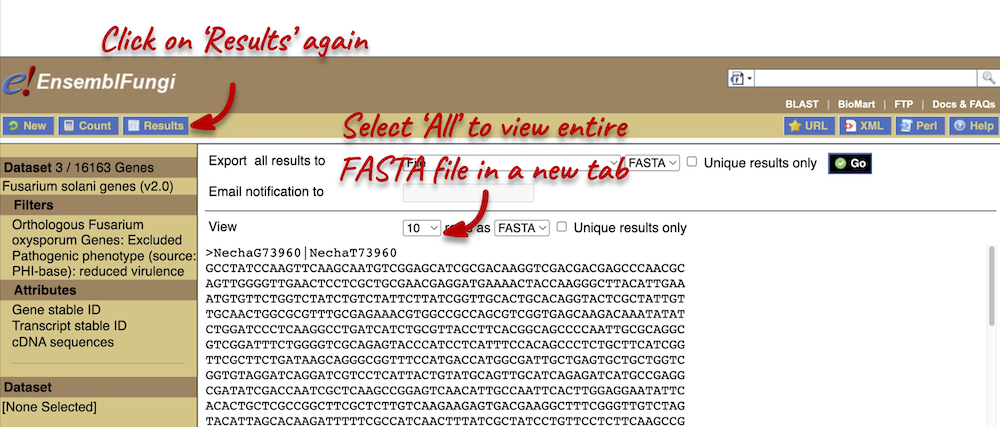
Also expand the HEADER INFORMATION tab and select Gene name. Click Results in the left-hand panel to see your sequences.
For more details on BioMart, have a look at this publication: Kinsella RJ, Kähäri A, Haider S, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database : the Journal of Biological Databases and Curation. 2011 ;2011:bar030. DOI: 10.1093/database/bar030. PMID: 21785142; PMCID: PMC3170168.






