Finding homologues and gene trees, demo
Let’s look at the homologues of the cow BRCA2 gene. Search for the gene and go to the Gene tab.
Click on Gene tree to display the current gene in the context of a phylogenetic tree used to determine orthologues and paralogues.
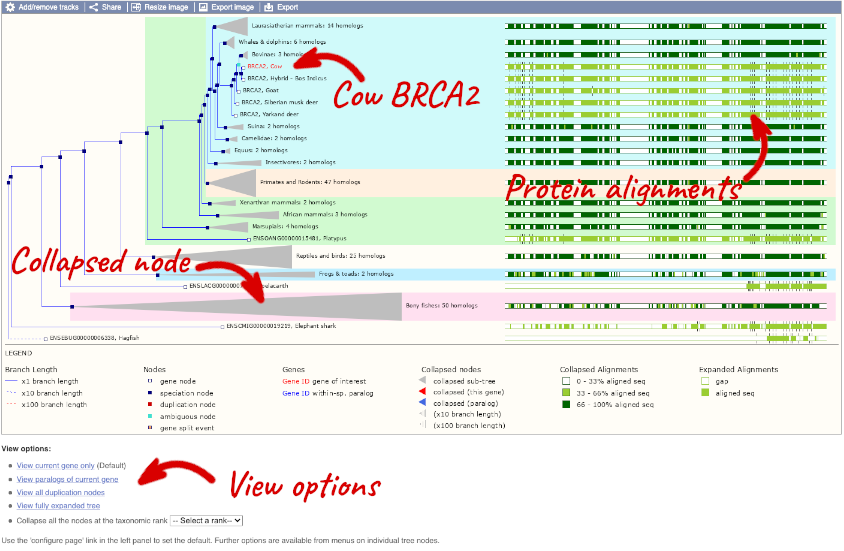
You can change the gene tree display by using the View options below the image, the Configure this page menu as well as menus for individual nodes, which you can open by clicking on the nodes. Grey funnels indicate collapsed nodes. You can expand them by clicking on the node and selecting expand this sub-tree from the pop-up menu.
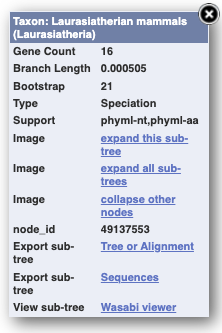
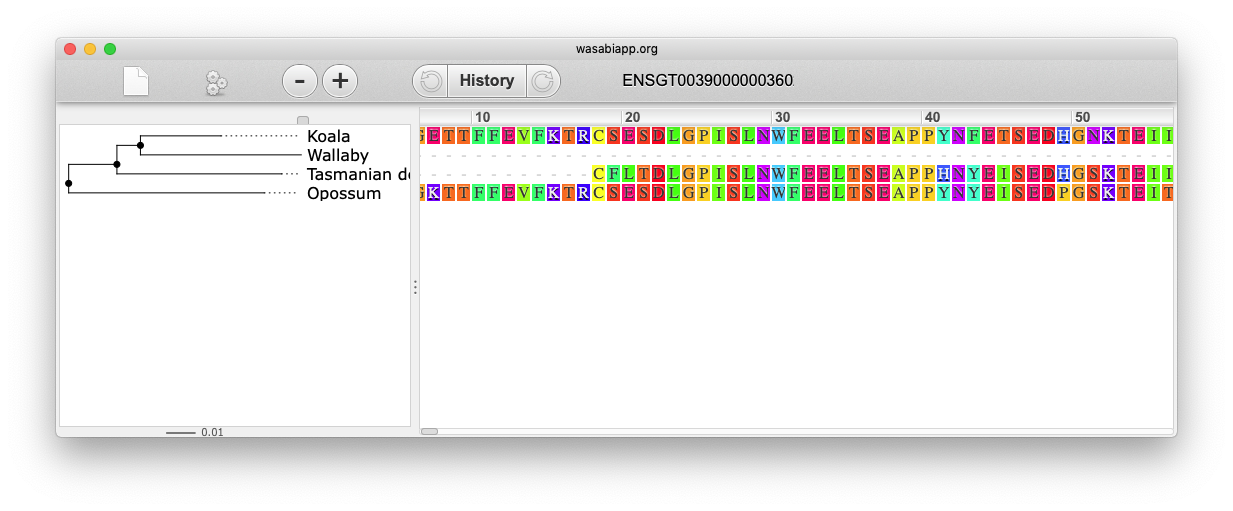
You can also view the alignment of the sub-tree by clicking on _Wasabi viewer _in the same menu, which will open a pop-up window:
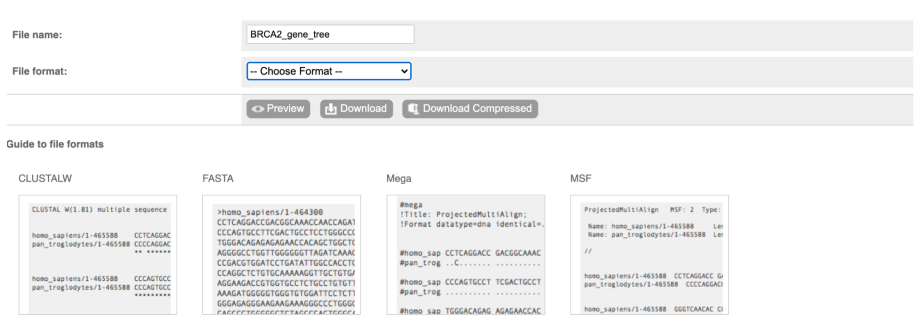
You can download the tree in a variety of formats. Click on the download icon in the bar at the top of the image to get a pop-up window where you can choose your format.

You can look at homologues in the Orthologues and Paralogues pages, which can be accessed from the left-hand menu. If there are no orthologues or paralogues, then the option will be greyed out. Paralogues is greyed out for BRCA2 indicating that there are no paralogues.
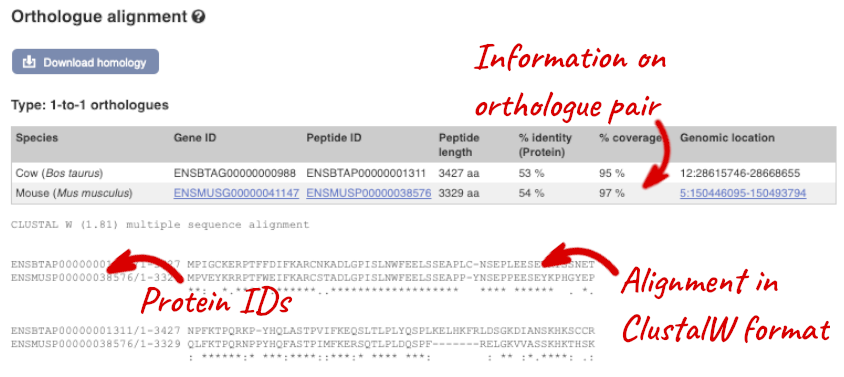
Click on Orthologues to see the available orthologues.
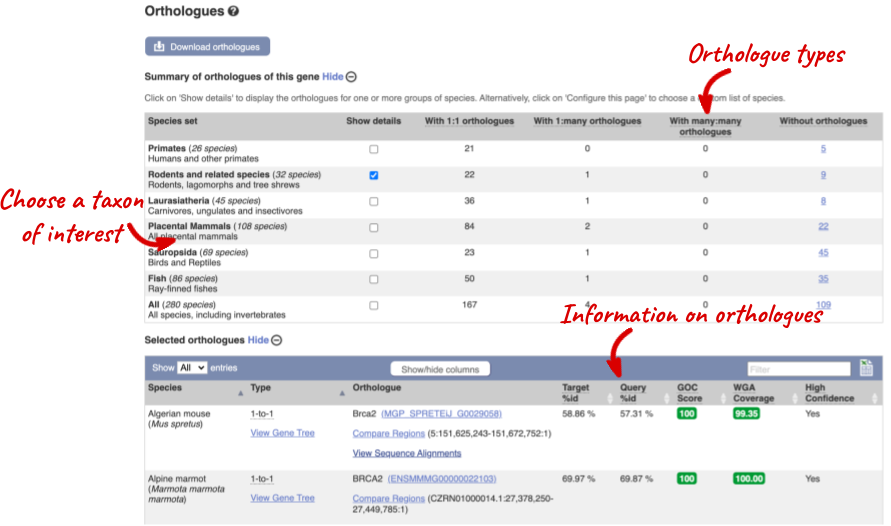
Choose to see only Rodent and related species orthologues by selecting the box. The table below now only shows details of these orthologues. Let’s look at mouse (Mus musculus).
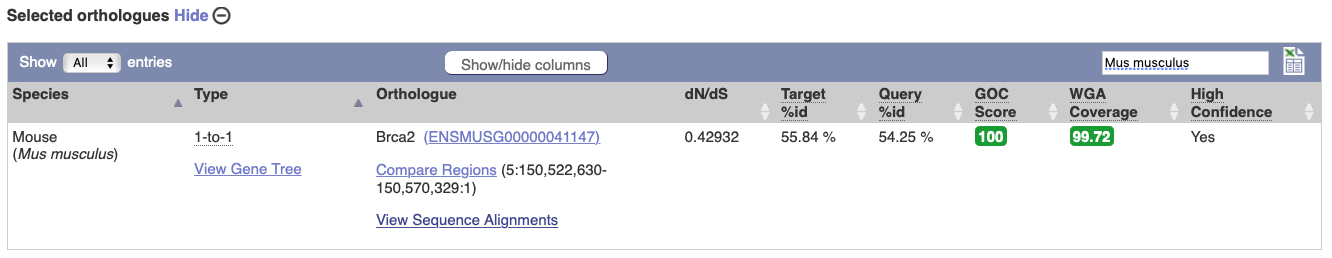
Links from the orthologue allow you to go to alignments of the orthologous proteins and cDNAs. Click on View Sequence Alignments, and then View Protein Alignment in the pop-up menu for the mouse orthologue.