Finding genetic variation data, demo
In any of the sequence views shown in the Gene and Transcript tabs, you can view variants on the sequence. You can do this by clicking on Configure this page from any of these views.
Let’s take a look at the Gene sequence view for MCM6 in chicken. Search for MCM6 and go to the Sequence view.
If you can’t see variants marked on this view, click on Configure this page and select Show variants: Yes and show links.
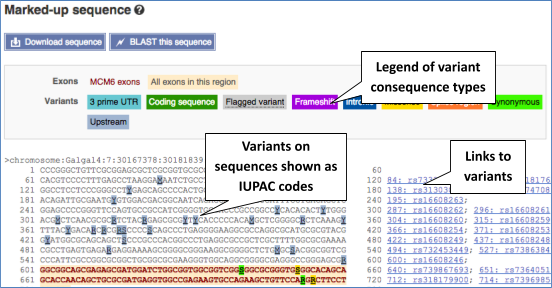
Find out more about a variant by clicking on it.
You can add variants to all other sequence views in the same way.
You can go to the Variation tab by clicking on the variant ID. For now, we’ll explore more ways of finding variants.
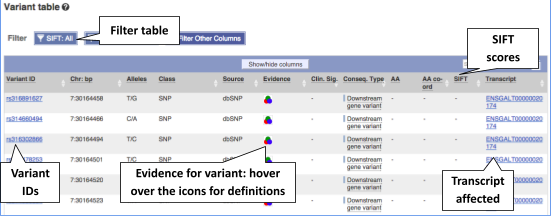
To view all the sequence variations in table form, click the Variant table link at the left of the gene tab.
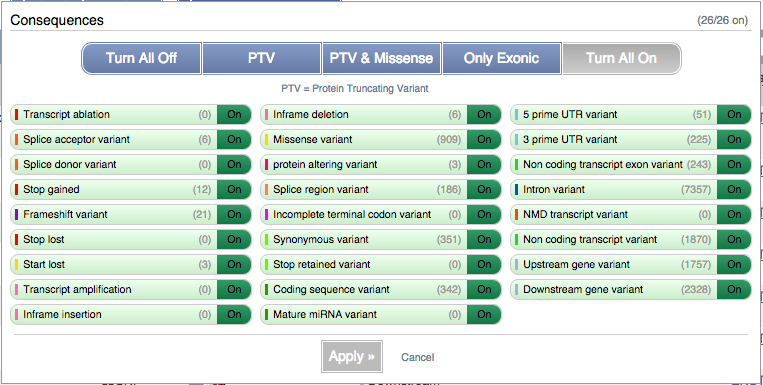
You can filter the table to only show the variants you’re interested in. For example, click on Consequences: All, then select the variant consequences you’re interested in.
You can also filter by SIFT, or click on Filter other columns for filtering by other columns such as Evidence or Class.
The table contains lots of information about the variants. You can click on the IDs here to go to the Variation tab too.
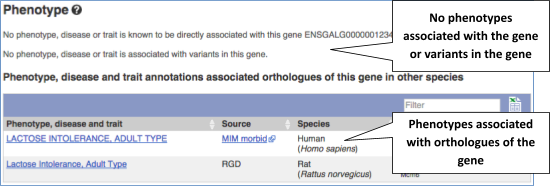
You can also see the phenotypes associated with a gene. Click on Phenotype in the left hand menu.
Let’s have a look at variants in the Location tab. Click on the Location tab in the top bar.
Click on Configure this page and open Variation from the left-hand menu.
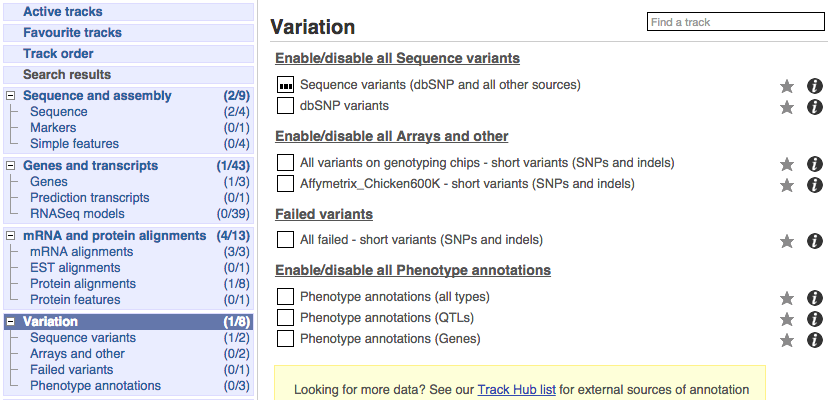
There are various options for turning on variants. You can turn on variants by source, by frequency, presence of a phenotype or by individual genome they were isolated from. You can also turn on QTLs, which cover a locus without being associated with a specific variant. Turn on the following variation tracks.
- All variants on genotyping chips - short variants (SNPs and indels)
- Phenotype annotations (QTLs)

Click on a variant to find out more information. It may be easier to see the individual variants if you zoom in.
Let’s have a look at a specific variant, which happens to fall within the MCM6 gene: rs14625781.
The easiest way to find this variant is if we put rs14625781 into the search box. Click through to open the Variation tab.
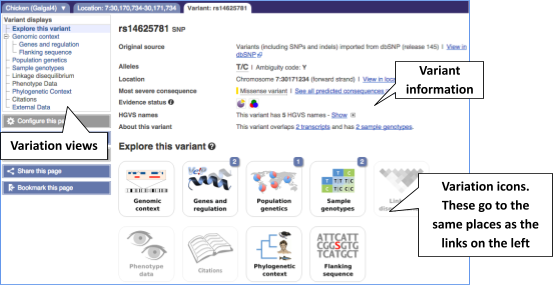
The icons show you what information is available for this variant. Click on Genes and regulation, or follow the link at the left.
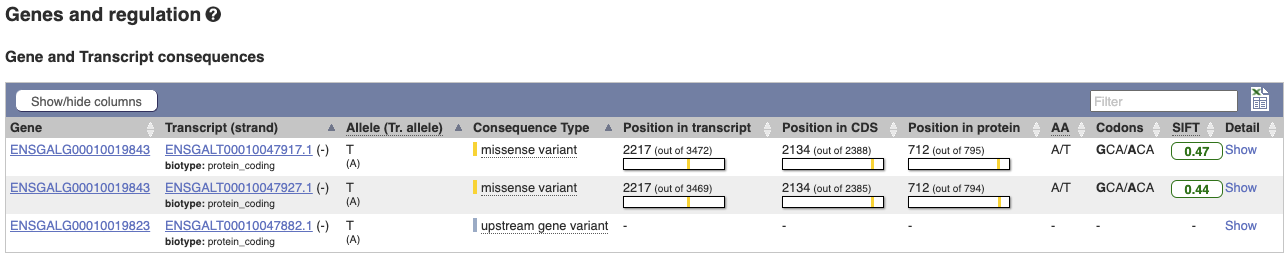
This variant is found in three transcripts of the MCM6 gene, and is missense in two. SIFT predicts that it is unlikely to affect protein function of either (Tolerated).
Let’s look at population genetics. Either click on Explore this variant in the left hand menu then click on the Population genetics icon, or click on Population genetics in the left-hand menu.
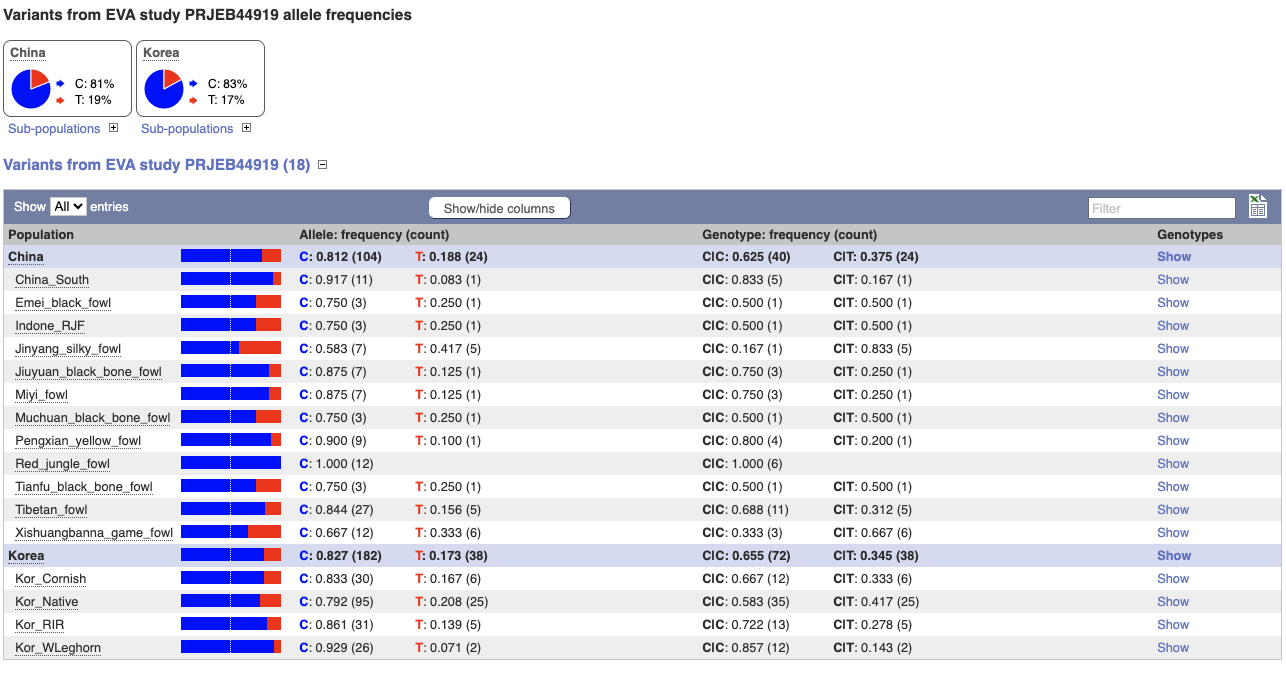
We can see data from EVA study PRJEB44919 showing the frequency of the alleles and genotypes. We can see what animals these genotypes were actually observed in by going to Sample genotypes.
Click on Phylogenetic context to see the variant in other species.
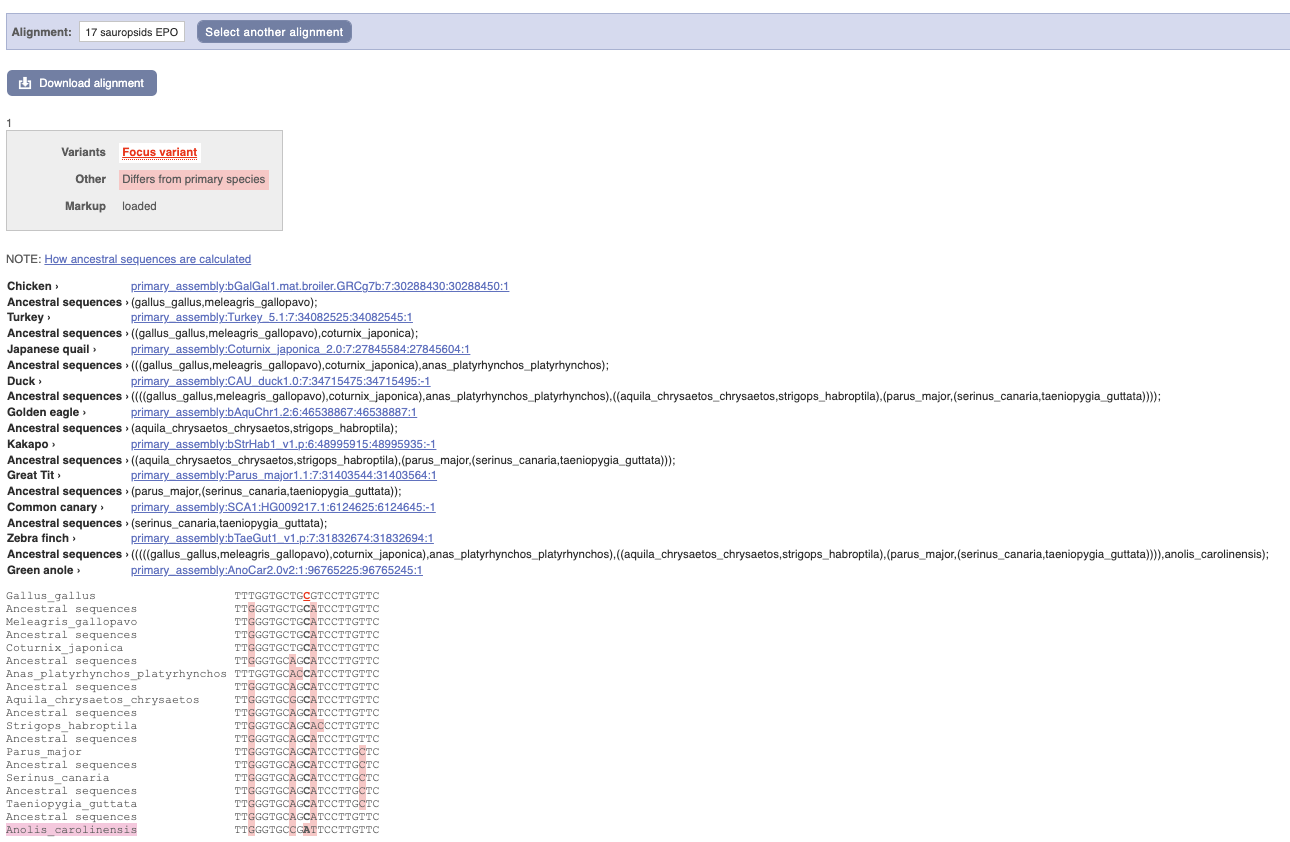
We can see that other birds also have the C alleles as a reference whereas Anolis_carolinensis has an A allele.






