Data export with BioMart
The BioMart tool can be used to export customized datasets directly from the Ensembl and Ensembl Genomes databases. It allows the extraction of the same data for a list of annotated features (genes, transcripts, variants, genomic regions).
Follow these instructions to guide you through BioMart to answer the following query:
We are interested in finding variants associated with snRNAs encoded on chromosome 1 of barley, H. vulgare. In particular, we want to discover:
- which snRNAs are encoded on chromosome 1H
- what strand they are transcribed from, and their genomic coordinates
- which variants are associated with the snRNA genes
- where they lie (strand and genomic coordinates)
- the variant alleles and their consequences
Click the BioMart link in the navigation bar, or go to http://plants.ensembl.org/biomart/martview/, to start:
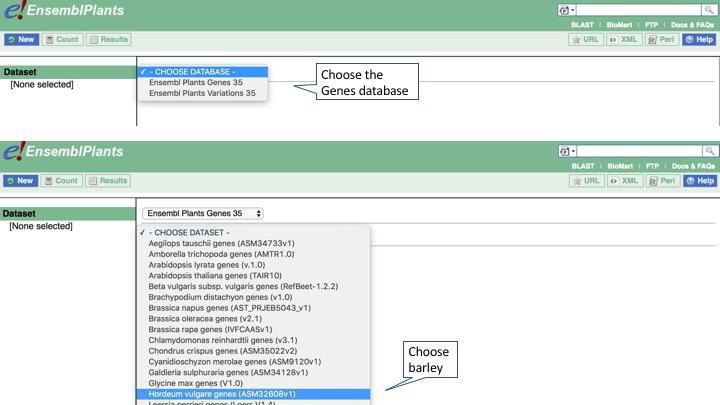
Now choose your dataset. Select Ensembl Plants Genes for the database, and _Hordeum vulgare genes_as the dataset.
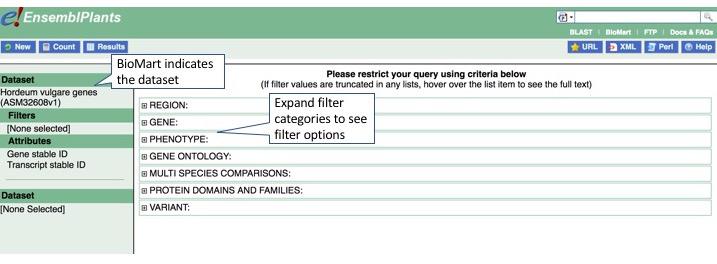
Next, we can apply filters. First, we want to filter on a genomic region, chromosome 1H. Click Filters, then expand Region. Select chr1H in the Chromosome/Scaffold box.
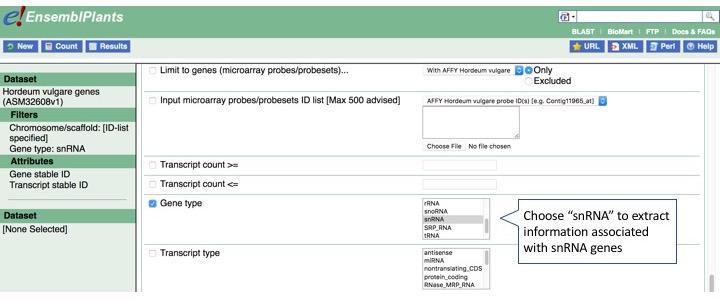
Next, we can apply our second filter to select only genes encoding snRNAs - the small nuclear RNAs that function as part of the spliceosome. Expand Gene to proceed, then in Gene type click snRNA.
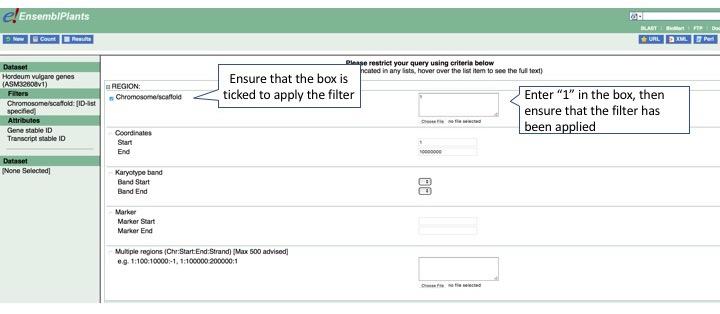
Check to make sure that both filters are applied. To determine the size of your filtered dataset, click the Count button just above the left-hand panel.
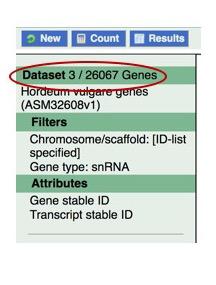
Now that we have filtered our dataset, we can apply Attributes. Start by clicking the Attributes link, on the left, to view possible options.
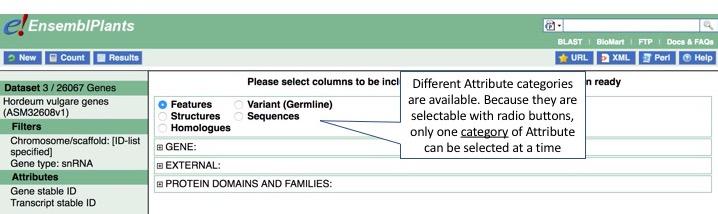
Features is selected by default. We are interested in Attributes associated with Variant (Germline); to proceed, click the button next to this option.
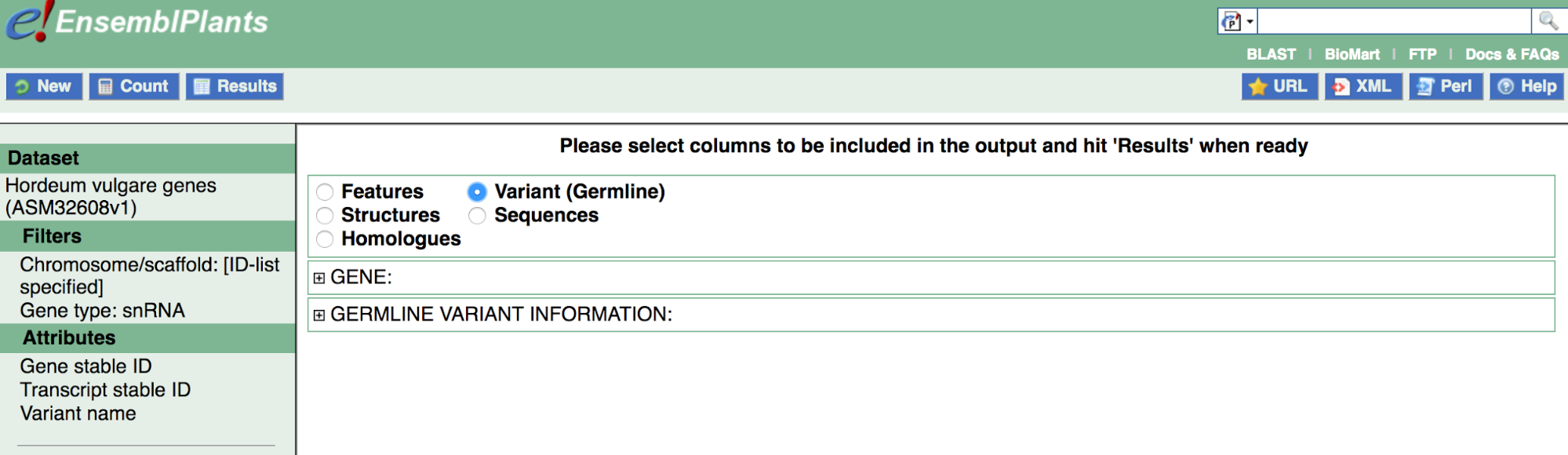
Now expand Gene and Germline Variant Information to see possible Attribute options. Note that a few options are pre-selected in each category; in Variant (Germline), Ensembl Gene stable ID, Ensembl Transcript stable ID and Variant name are pre-selected.
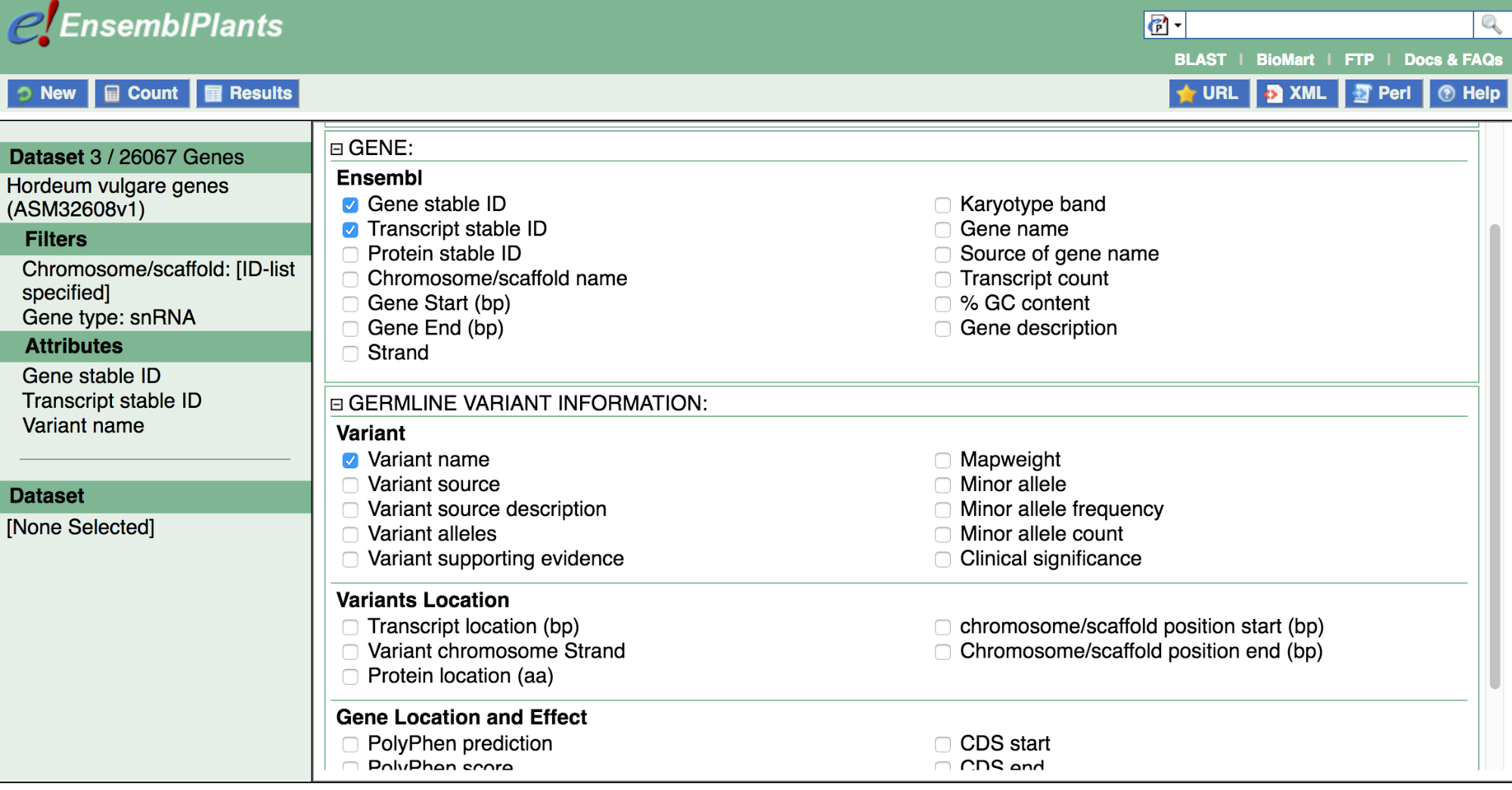
Results columns will be printed in the Results table in the order in which the Attributes are selected. If you would like to re-order Attributes, you can simply untick and re-tick them.
Let’s select Attributes now. We are interested in applying Attributes that will help us determine which snRNAs are encoded on barley chromosome 1, as well as their genomic coordinates; and also the variants that have been associated with them, their alleles, and their genomic coordinates.
We’ll select the Attributes shown in the two panels below:

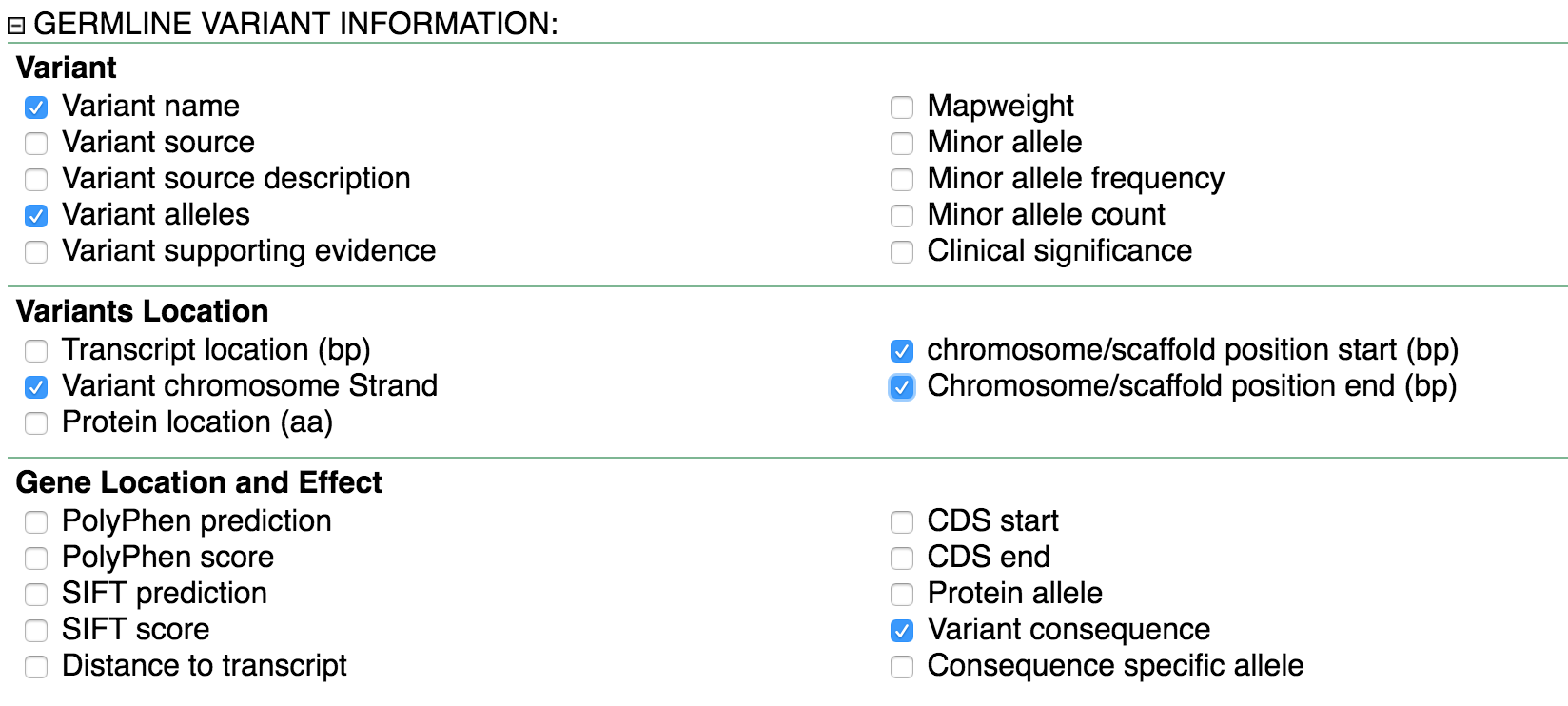
Our full query is as follows:
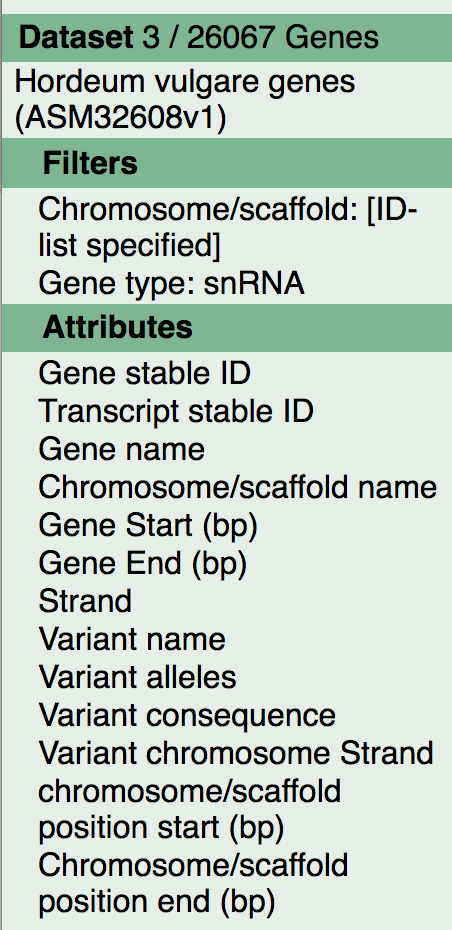
We can now generate our results. Simply hit the Results button above the left-hand panel to print out a preview of your results.
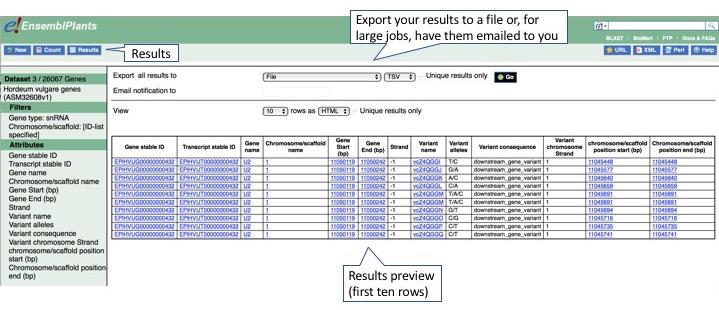
Use the dropdown menus to export your results, or to view an expanded table in the browser.






