Finding genomics data in Ensembl Beta, Demo
This is the homepage for the new Ensembl, which is currently released as a beta version. Over time additional species and functionality will be added, but for now let’s start by choosing a species using the “Species selector”.
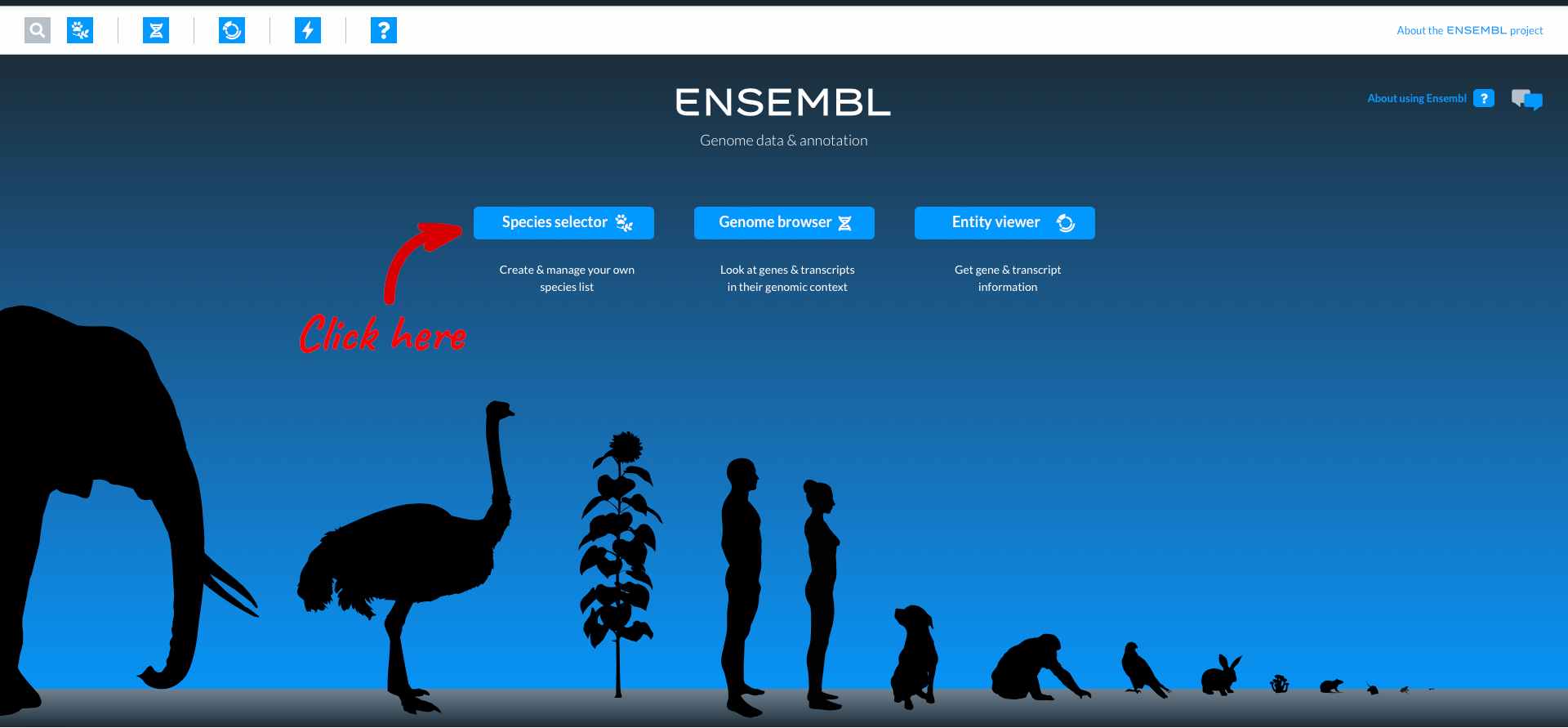
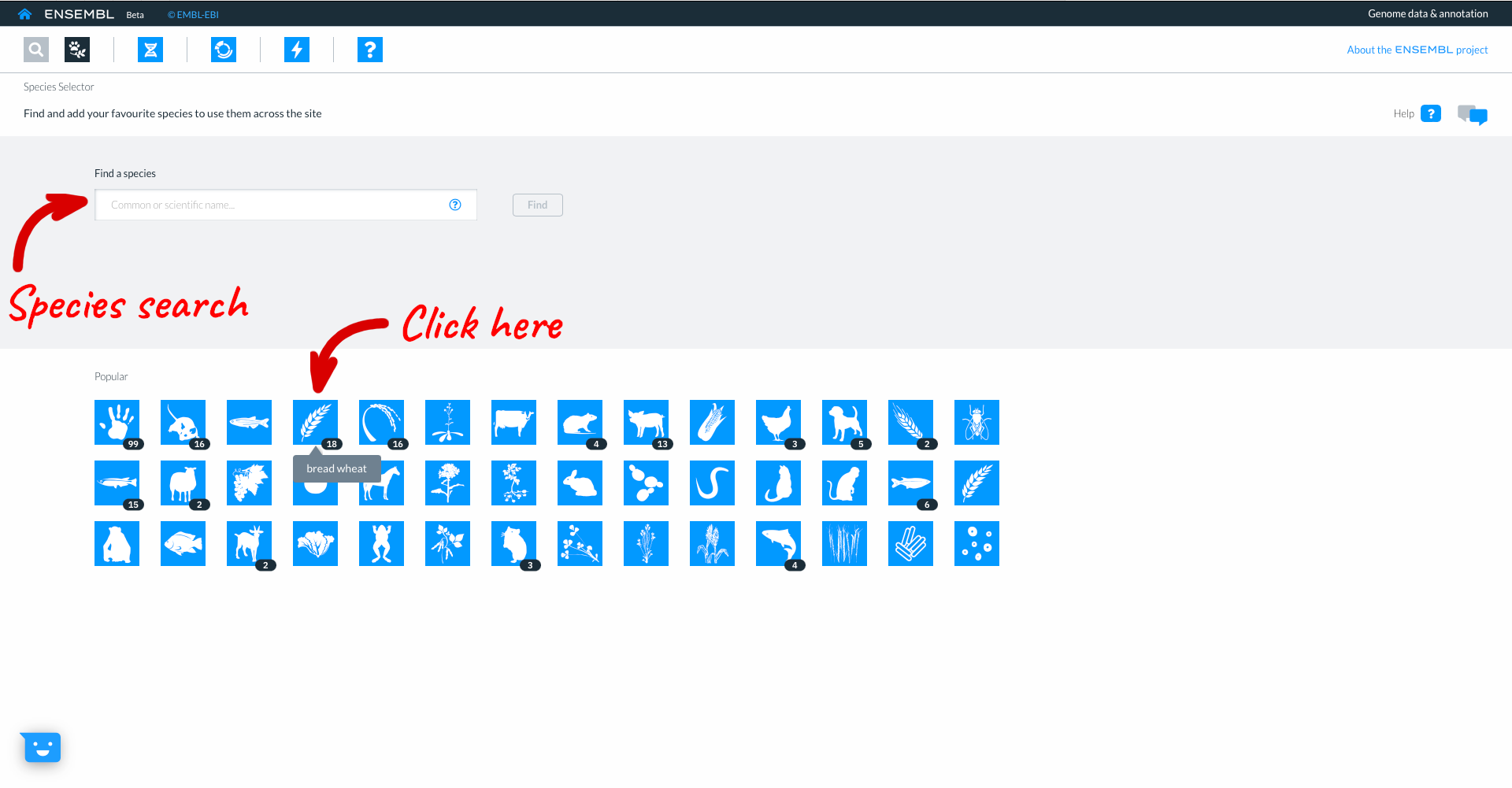
Here you can see icons for the most popular species in Ensembl, comprising vertebrates, invertebrates, plants, fungi and microbes. You can select a species to work with by clicking on the icon. The numbers in black lozenges at the bottom corner of species icons show where we hold more than one assembly for that species. You can also select species by typing into the species search box. We are going to work with wheat, so click on the wheat icon.
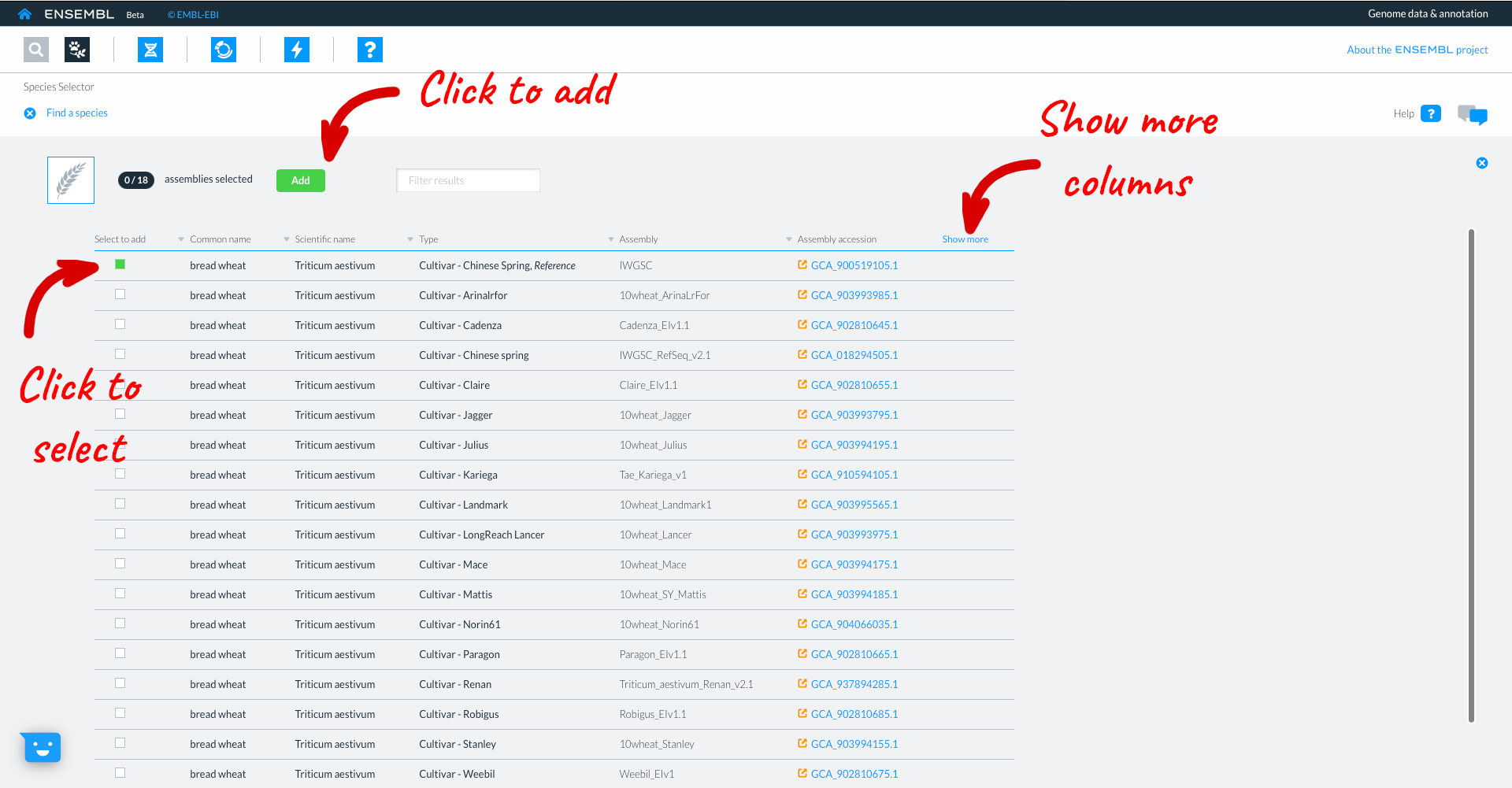
This takes you to the list of available wheat genomes currently available in Beta Ensembl. The table provides information such as common name, scientific name, and information about the assembly. Clicking ‘show more’ reveals additional columns with information relating to the assembly quality and additional data types associated with that assembly. You can select genomes to work with by clicking the square button to the left of each genome. We will select the wheat reference IWGSC assembly, then click “Add”.
Clicking on the genome name in the top panel will take you to the species page for that genome where more detailed information and statistics relating to that assembly are displayed.
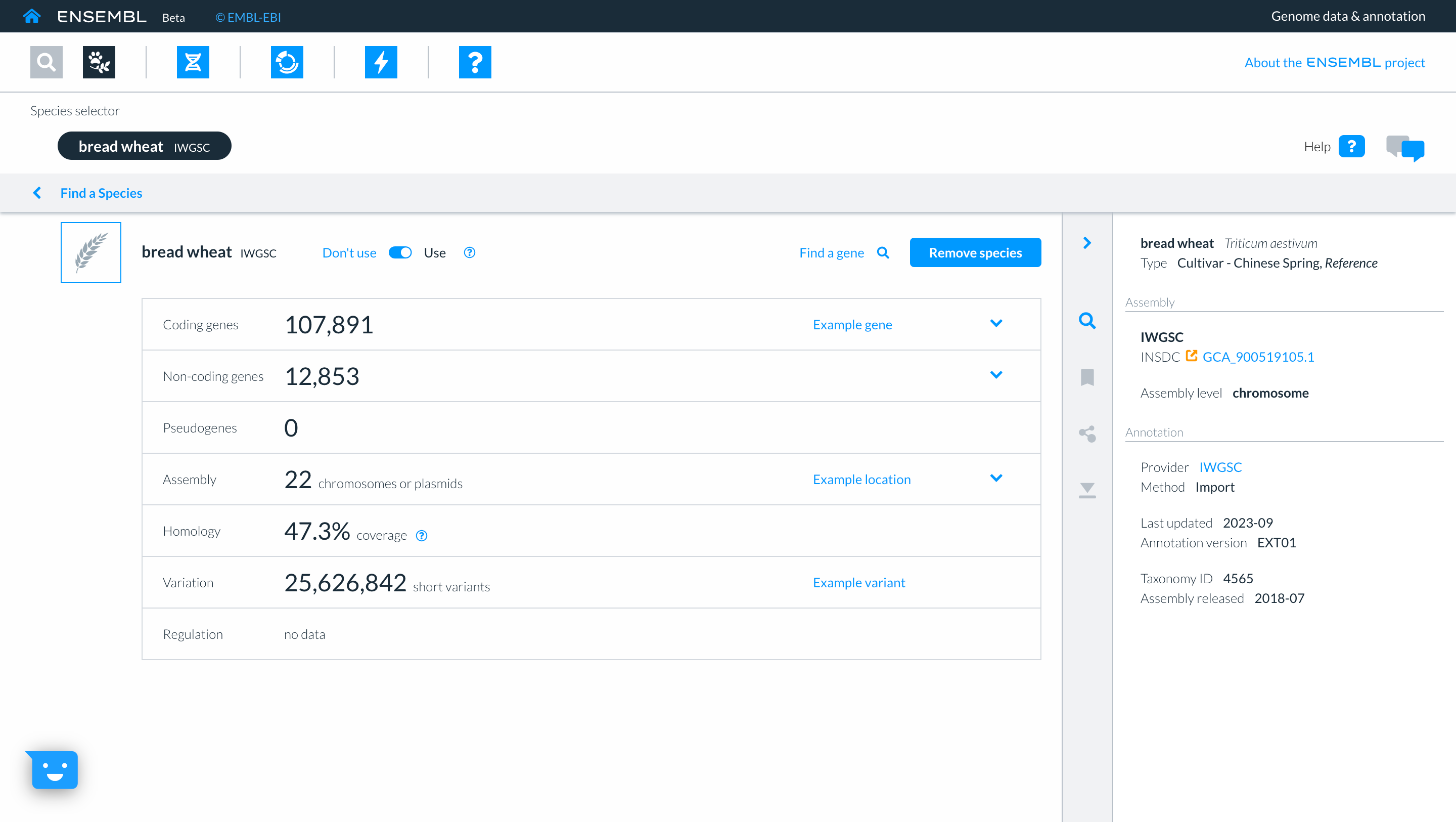
Returning to the species selector by clicking on ‘Find a species’, you can now see the genomes which you have selected in the upper part of the screen. Here too you have icons to access other parts of Ensembl. For now we will click on the ‘Genome Browser’ icon and then searching for gene “TraesCS3D02G273600”.
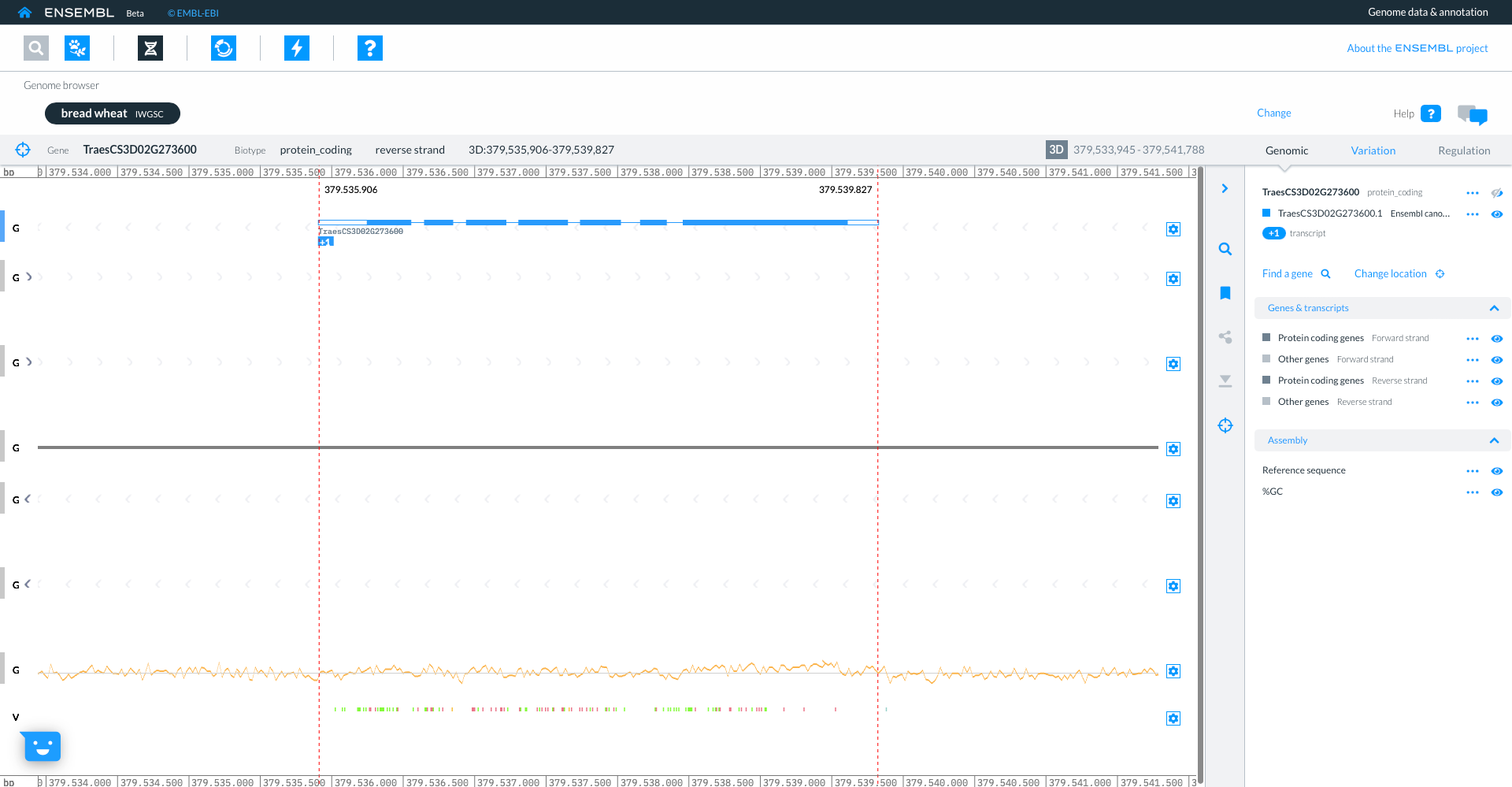
The genome browser shows gene model TraesCS3D02G273600 with exons in blue boxes, introns as lines and UTR as open boxes. The wiggle plot shows the GC content, while the bottom track show variants coloured by consequence. The right hand panel gives more information about the transcripts in the focus gene.
If you click on the transcript model a pop-up appears with an overview of the gene and Download options for the different sequences e.g. genomic, cDNA, protein or CDS. 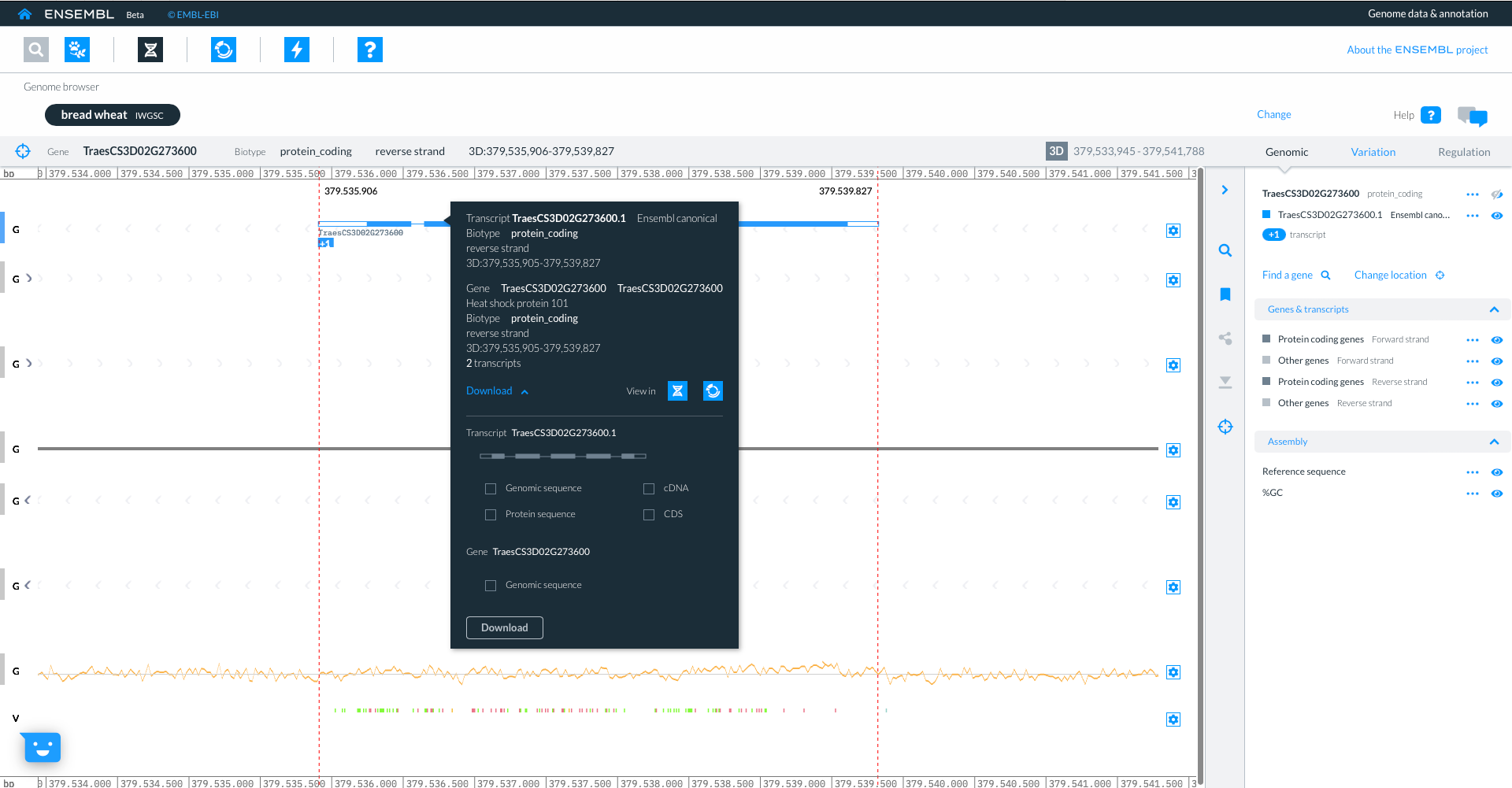
If you zoom in you can focus on the individual variants, with further details shown in the right hand panel
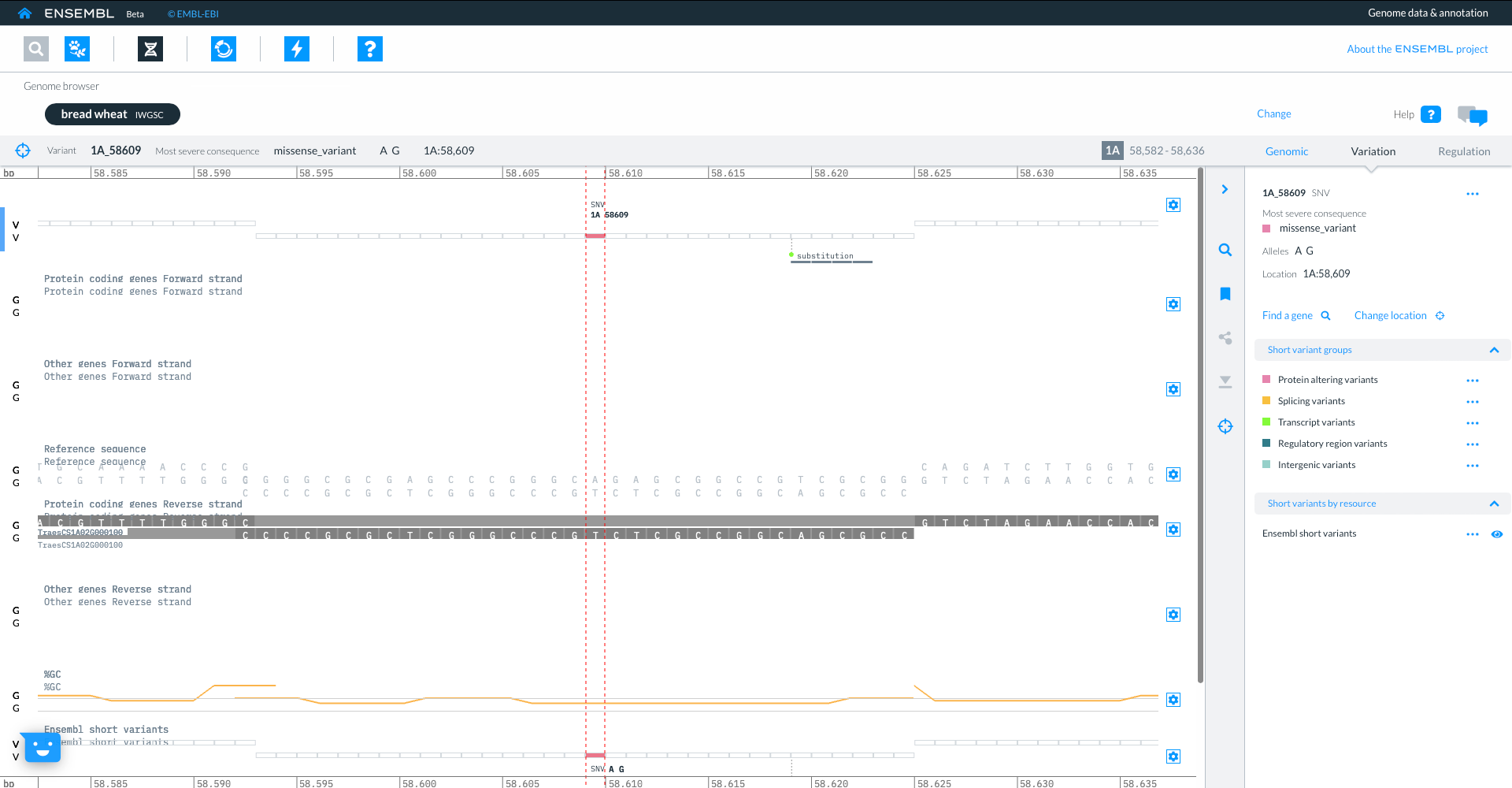
Using the icons in the top panel we can also go to the entity viewer, where we can find more details about the entities in question. Here we are focused on the same gene TraesCS3D02G273600 and we can view and download sequence from the individual transcripts.
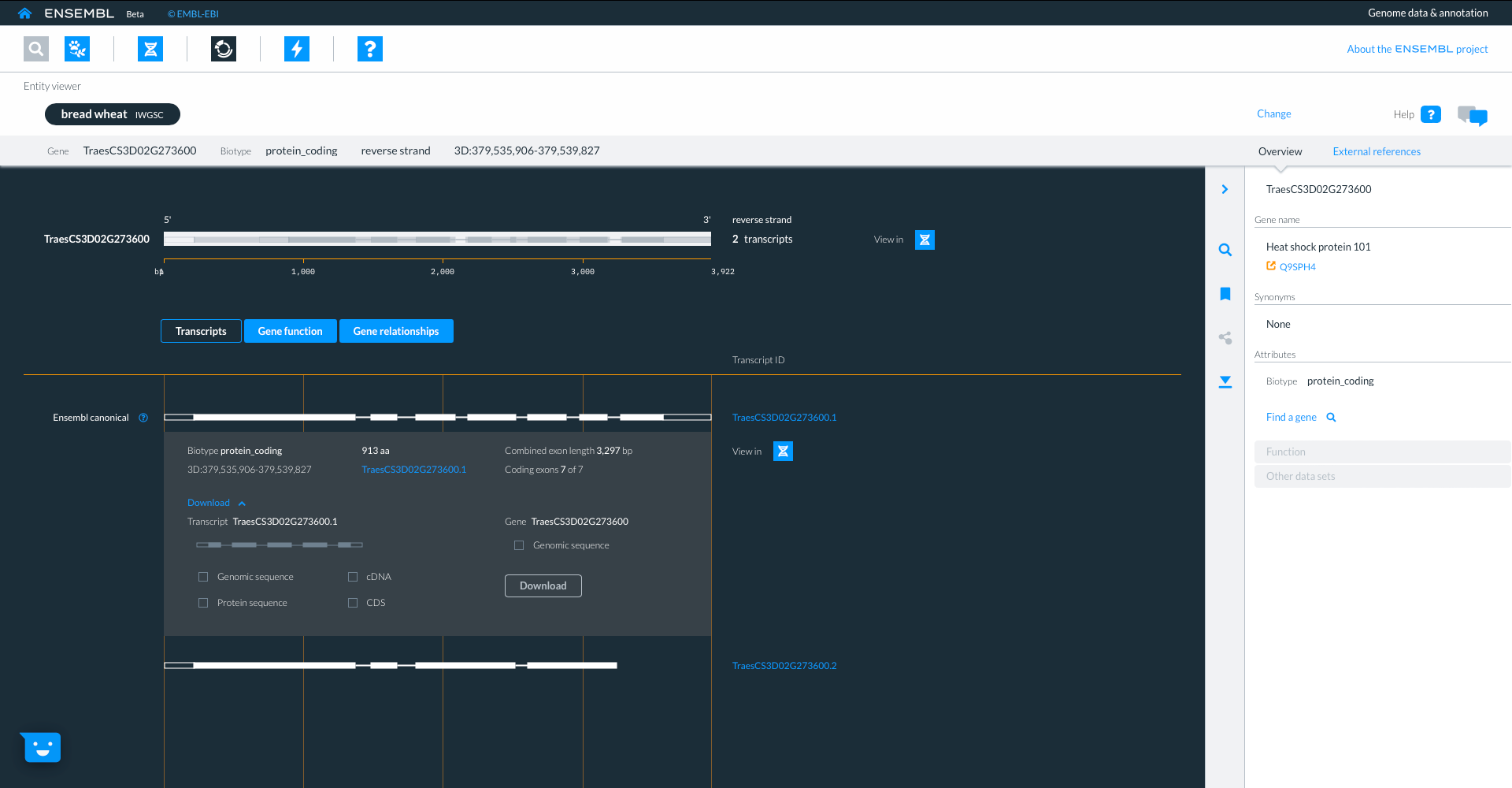
We can also look at the ‘Gene function’ tab where we see information about protein domains, with links to Uniprot and Interpro. We can also see information on variants, phenotypes, gene expression and pathway information where available.
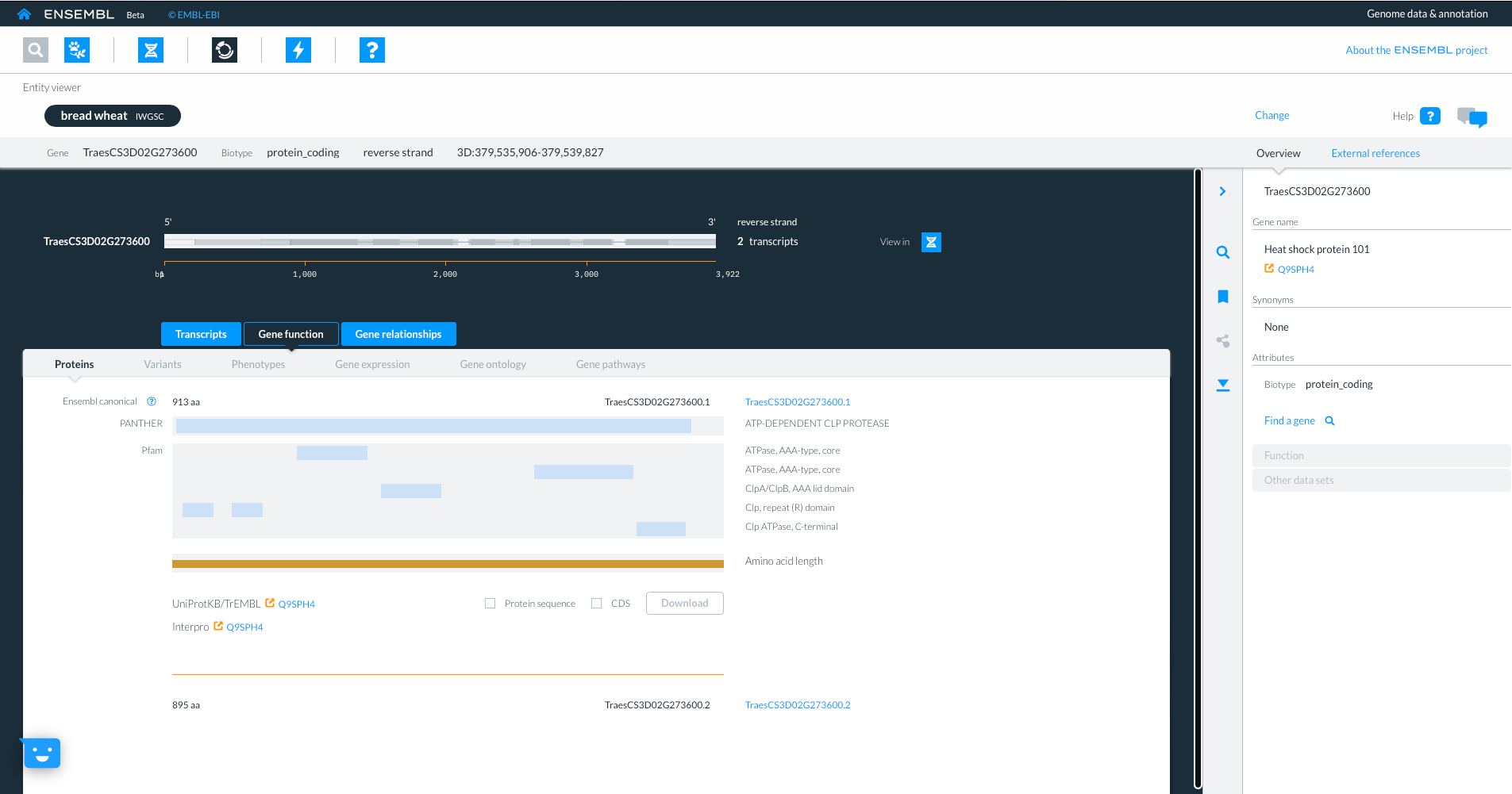
Selecting the third tab “Gene relationships” shows homology information to other ensembl species, with details about sequence similarity and how the hit was identified. By selecting links to homologue gene IDs we can jump to the homologues gene in another species. We will select gene “TRITD3Bv1G162500” from Triticum turgidum ssp. Durum.
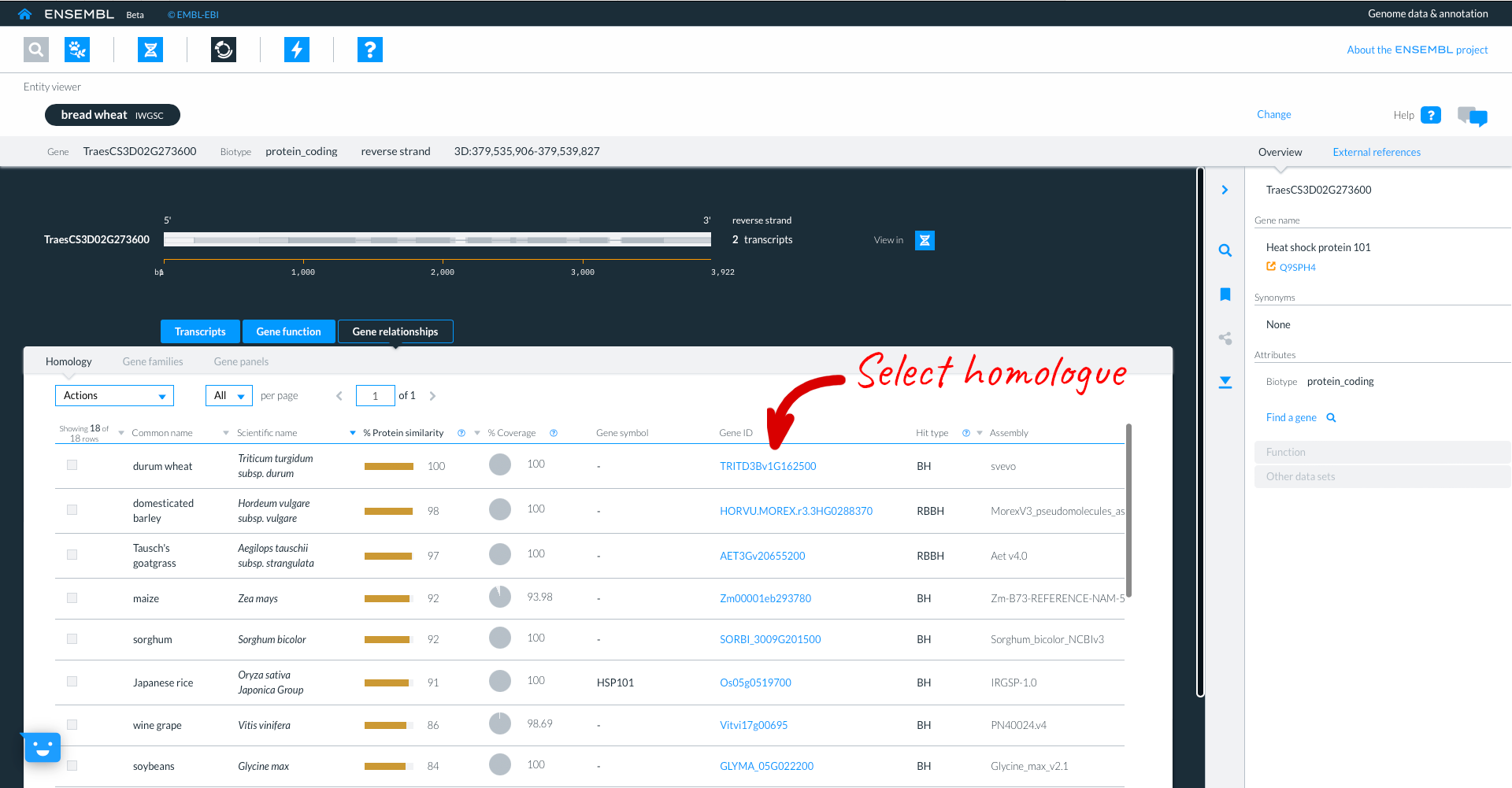
Here we see the Durum gene, in the entity viewer. The focus species is coloured in black, but the original species (in our case Bread wheat T. aestivum remains listed to allow for easy switching between the two).
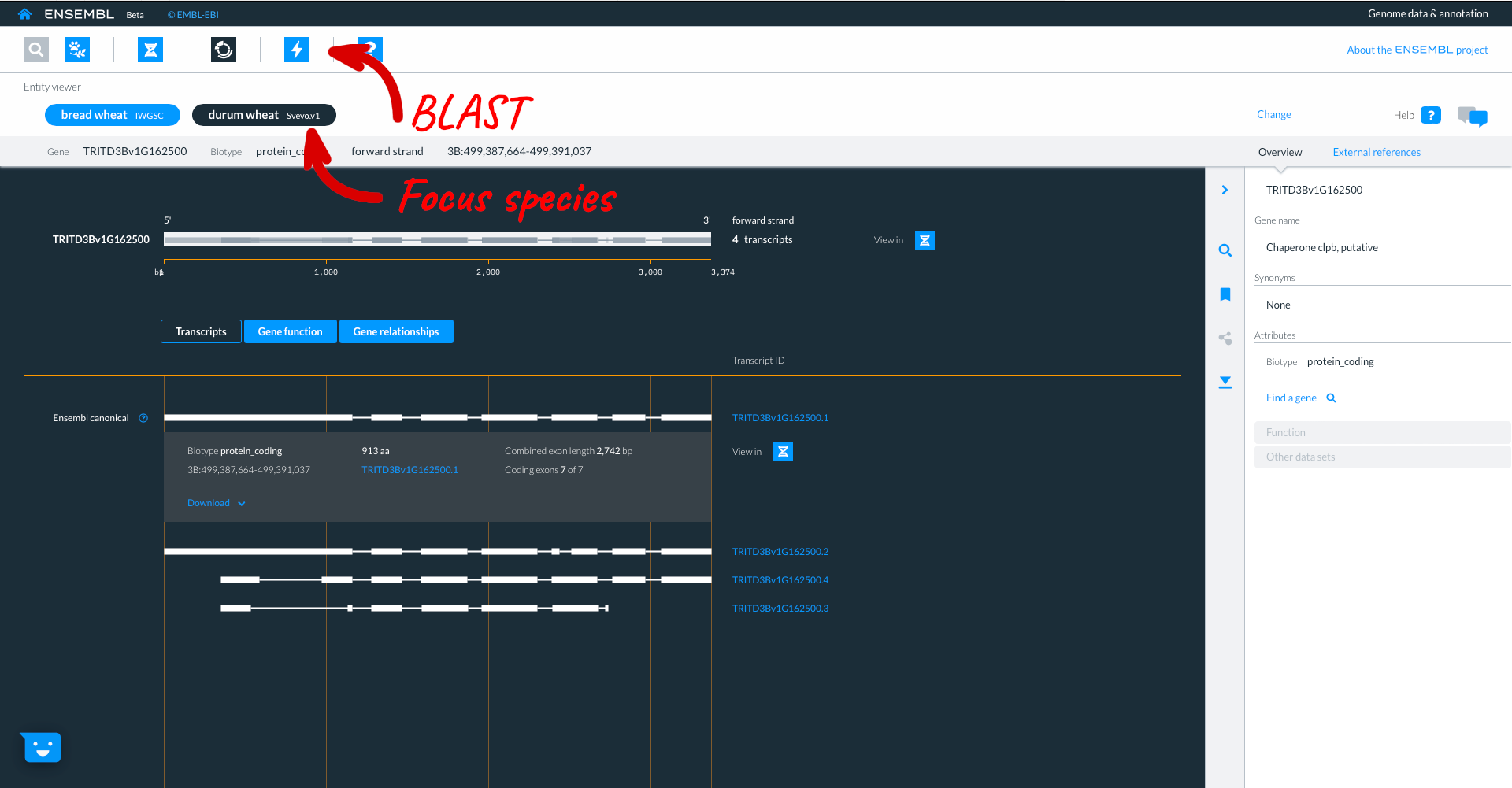
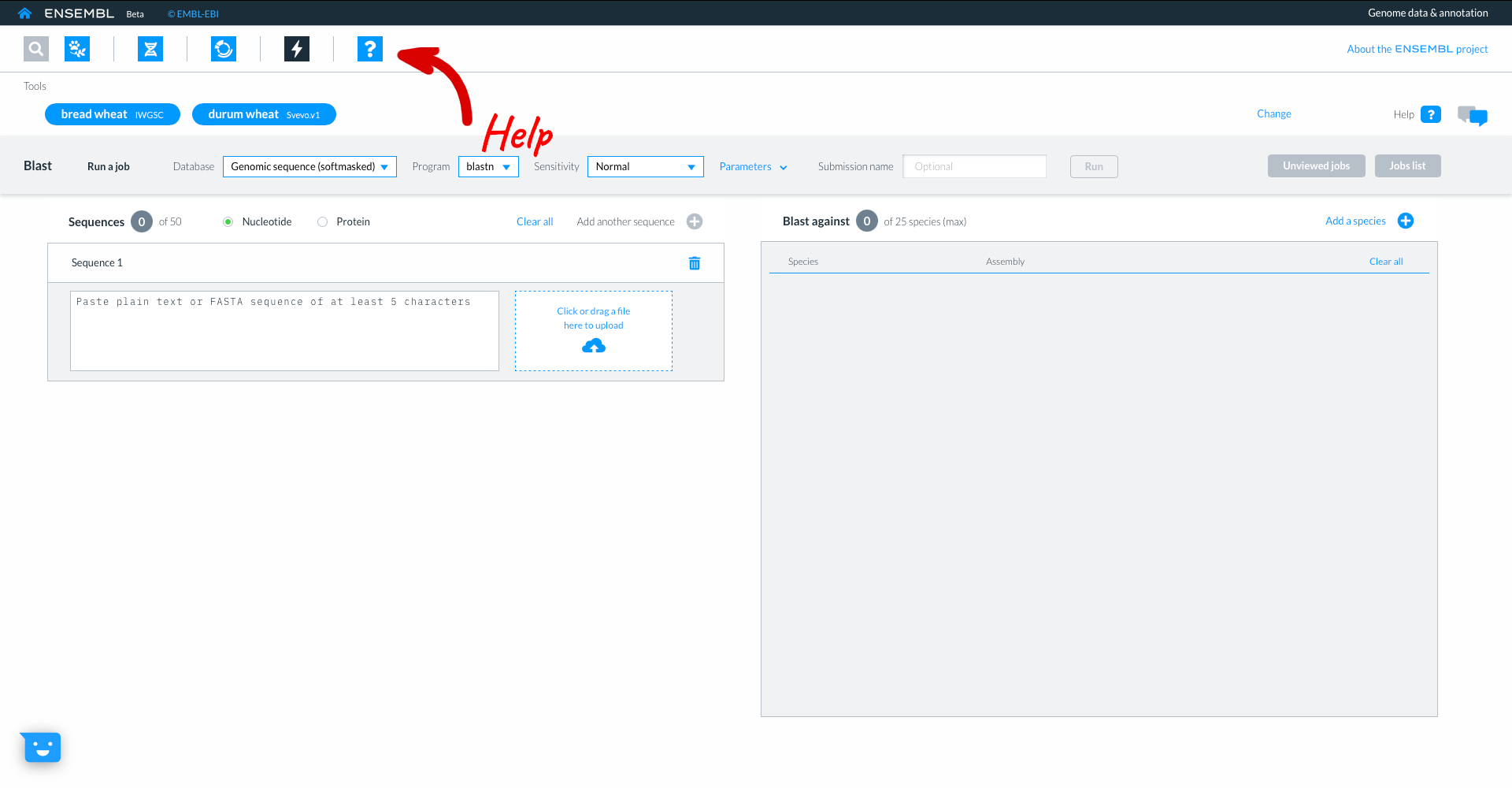
Beta Ensembl has BLAST functionality which can be accessed by clicking on the lightning icon in the top panel. Where you can currently BLAST up to 50 sequences against up to 25 species.
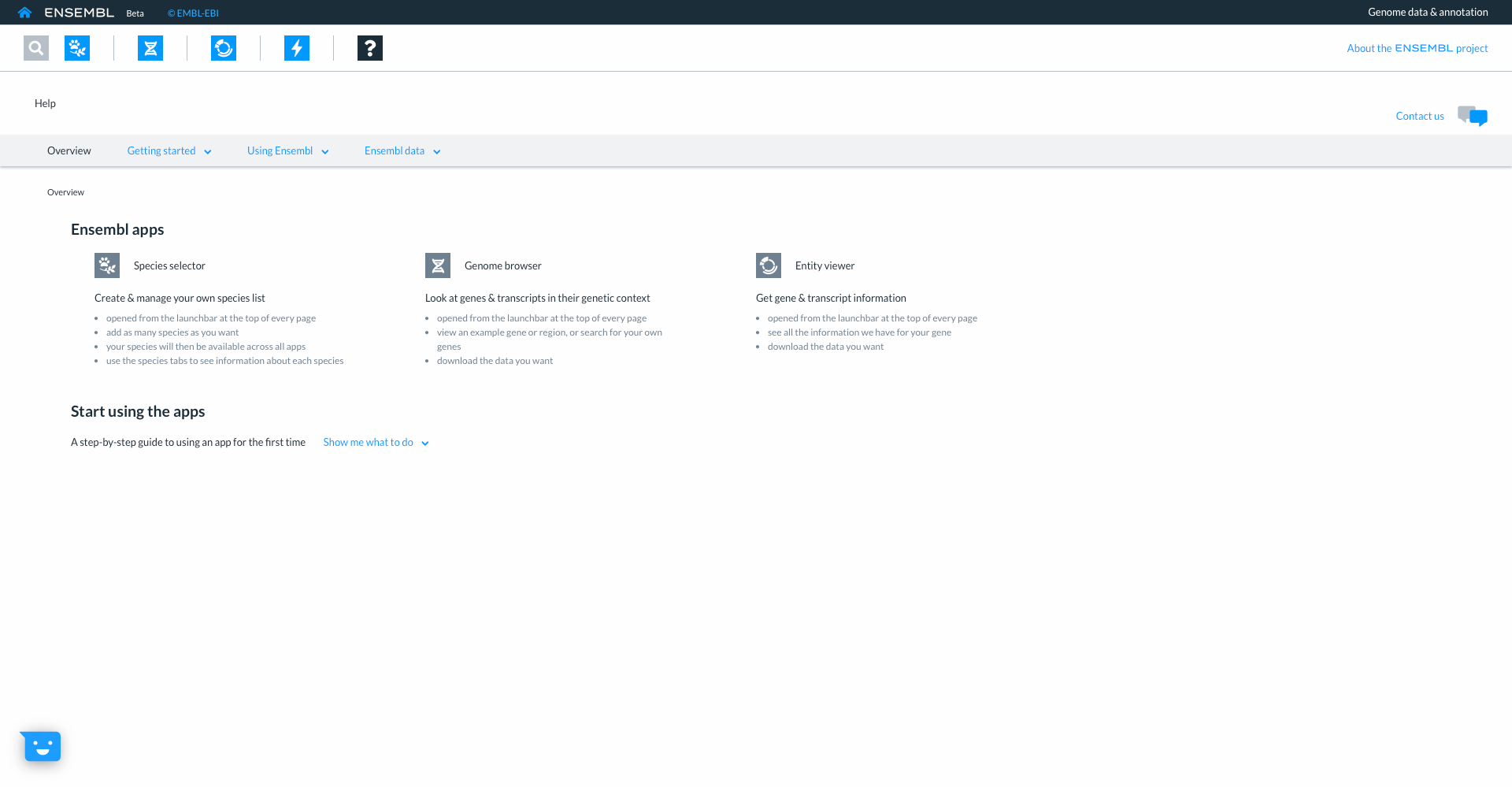
The final icon with the ‘?’ links to our help and documentation where you can learn more about the different Ensembl apps.
On every page you will also find a speech bubble icon where you can quickly feedback on your experience when using the site.
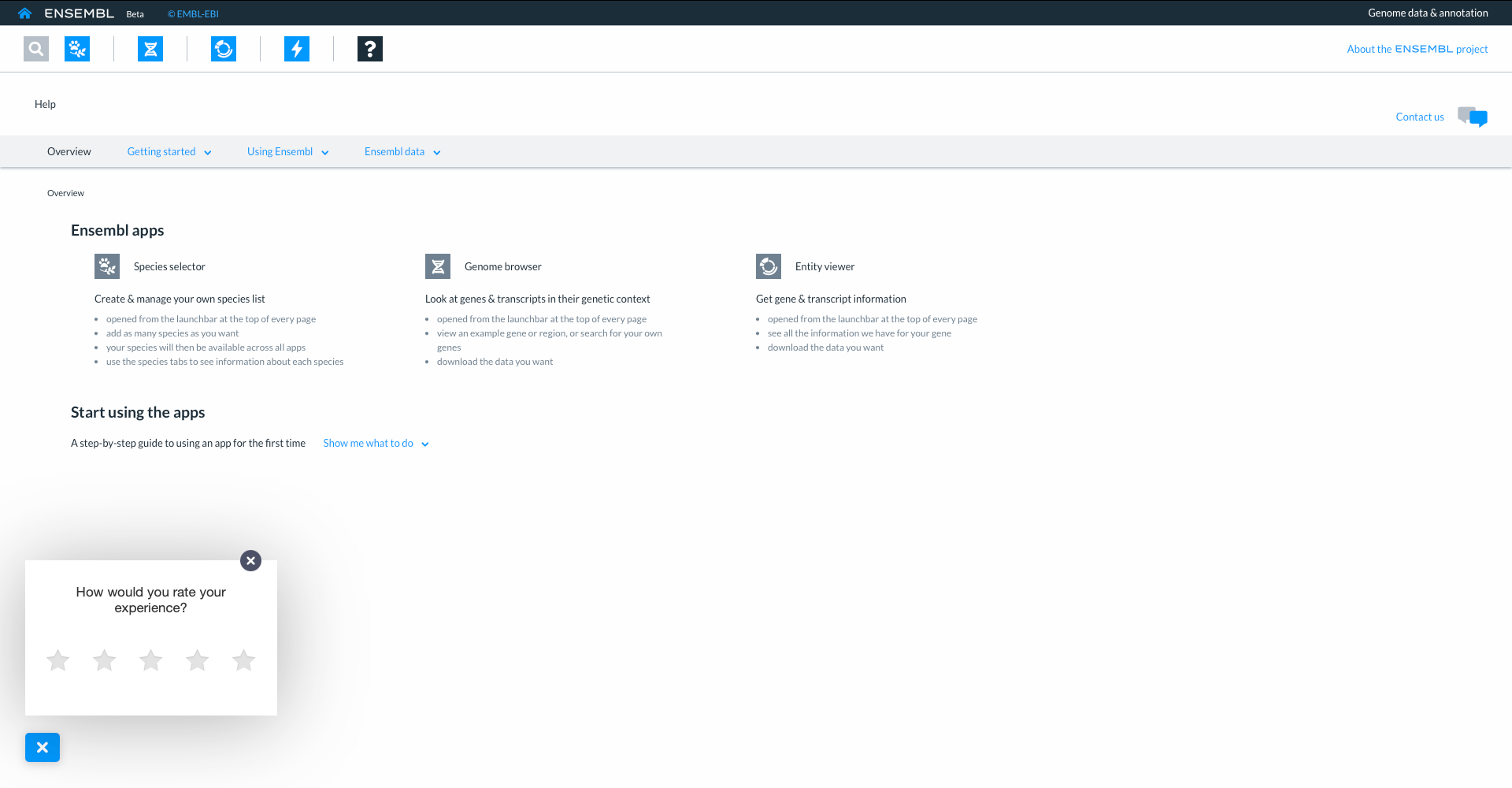
You will also find a ‘contact us’ icon on the top right of every page where you can get In touch to leave more detailed feedback or ask questions. Links to the blog, our social media and mailing lists are also provided.