Finding genes and transcripts in Ensembl, Demo
Demo: The gene tab
If you click on any one of the transcripts in the Region in detail image, a pop-up menu will appear, allowing you to jump directly to that gene or transcript.
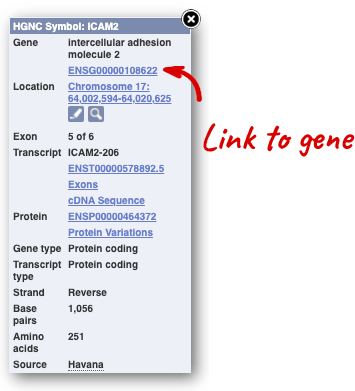
Another way to go to a gene of interest is to search directly for it.
We’re going to look at the fungus Zymoseptoria tritici MK1 gene.
From fungi.ensembl.org, type MK1 into the Search for a gene search bar and click the Go button. You will get a list of hits with the Puccinia sorghi gene at the top.
Where you search for something without specifying the species, or where the ID is not restricted to a single species, you can restrict your query to species using the drop down at the top.
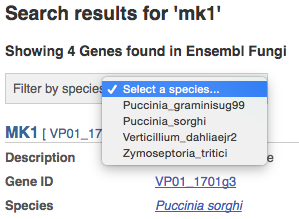
Click on the Zymoseptoria_tritici to narrow down, then on MK1 in the results. The Gene tab should open:
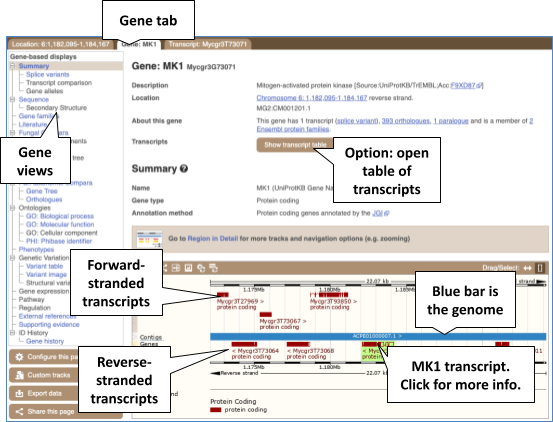
Let’s walk through some of the links in the left hand navigation column. How can we view the genomic sequence? Click Sequence at the left of the page.
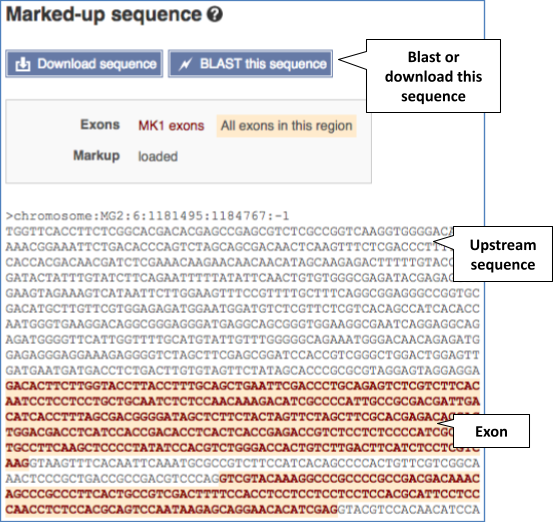
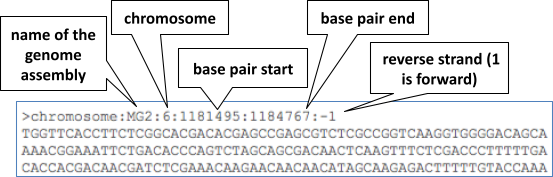
The sequence is shown in FASTA format. Take a look at the FASTA header:
Exons are highlighted within the genomic sequence. Variants can be added with the Configure this page link found at the left. Click on it now.
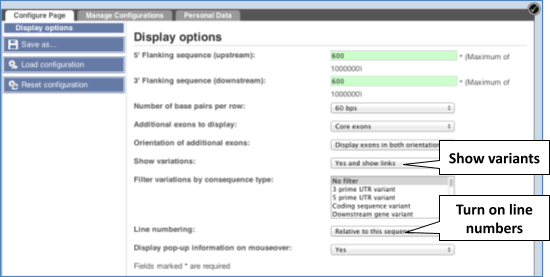
Once you have selected changes (in this example, Show variants and Line numbering) click at the top right.

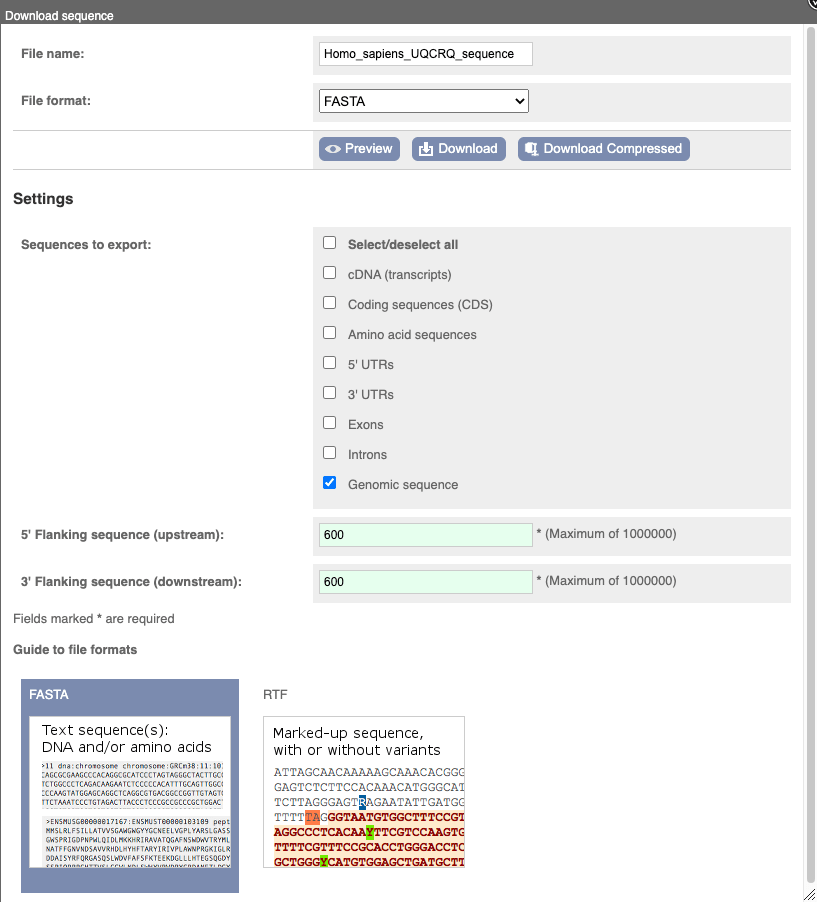
You can download this sequence by clicking in the Download sequence button above the sequence:

This will open a dialogue box that allows you to pick between plain FASTA sequence, or s_equence in RTF_, which includes all the coloured annotations and can be opened in a word processor. This button is available for all sequence views.
To find out what the protein does, have a look at GO terms from the Gene Ontology consortium (http://www.geneontology.org). There are three pages of GO terms, representing the three divisions in GO: Biological process (what the protein does), Cellular component (where the protein is) and Molecular function (how it does it). Click on GO: Biological process to see an example of the GO pages.

Since this gene has data from PHI-base, we can explore this in the Gene tab too. Click on PHI: Phibase identifier in the left-hand menu.
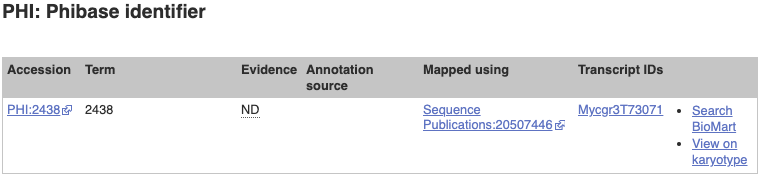
This page is very similar to the GO pages.
Demo: The transcript tab
Many genes have multiple transcripts which can be seen in the transcript table. Click on Show transcript table at the top.

We can go to the transcript tab either by clicking on the transcript ID, Mycgr3T73071, in the table, or on the transcript tab, Transcript: Mycgr3T73071, at the top.
You are now in the Transcript tab. The left hand navigation column provides several options for the transcript.
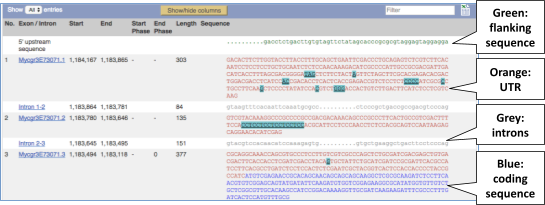
Click on the _Exons _link.
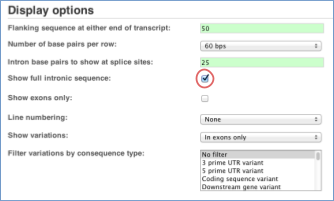
You may want to change the display (for example, to show more flanking sequence, or to show full introns). In order to do so click on Configure this page and change the display options accordingly.
Now click on the_ cDNA_ link to see the spliced transcript sequence.
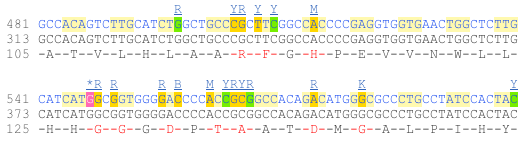
UnTranslated Regions (UTRs) are highlighted in dark yellow, codons are highlighted in light yellow, and exon sequence is shown in black or blue letters to show exon divides. Sequence variants are represented by highlighted nucleotides and clickable IUPAC codes are above the sequence.
Next, follow the General identifiers link at the left.
This page shows information from other databases such as RefSeq, UniProtKB, CCDS and others, that match to the Ensembl transcript and protein.
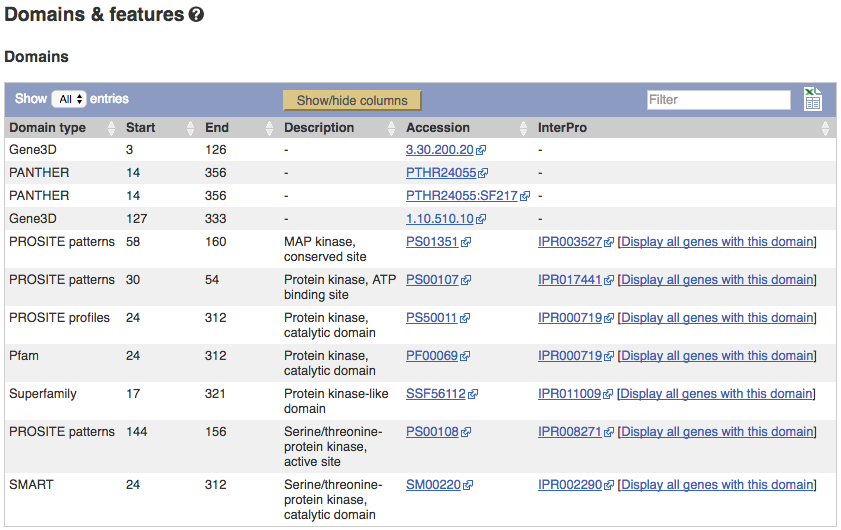
Now click on Protein summary to view domains from Pfam, PROSITE, Superfamily, InterPro, and more.

Clicking on Domains & features _shows a table of this information.