Filter Events by Year
Webinar: Introduction to the Ensembl Bacteria genome browser
Course Details
- Lead Trainer
- Louisse Paola Mirabueno
- Associate Trainer(s)
- Event Date
- 2024-04-24
- Location
- Virtual
- Description
- We will explore Ensembl Bacteria to learn more about the latest data and where it came from, view annotation of molecular interactions involving bacterial genes and comparative genomics data involving key species
Demos and exercises
Region in detail and Genes
Region in Detail view
Start at the Ensembl Bacteria homepage, bacteria.ensembl.org. Search for your species of interest either by using the search box, or opening the full list of species by clicking View full list of all Ensembl Bacteria species underneath the search box.
Enter Escherichia coli str. K-12 substr. MG1655 (GCA_000005845) in the search box. Enter Chromosome:3144663-3157453 into the species-specific search box:
Press Enter or click Go to jump directly to the Region in detail page.
Click on the  button to view page-specific help. The help pages provide text, labelled images and, in some cases, help videos to describe what you can see on the page and how to interact with it.
button to view page-specific help. The help pages provide text, labelled images and, in some cases, help videos to describe what you can see on the page and how to interact with it.
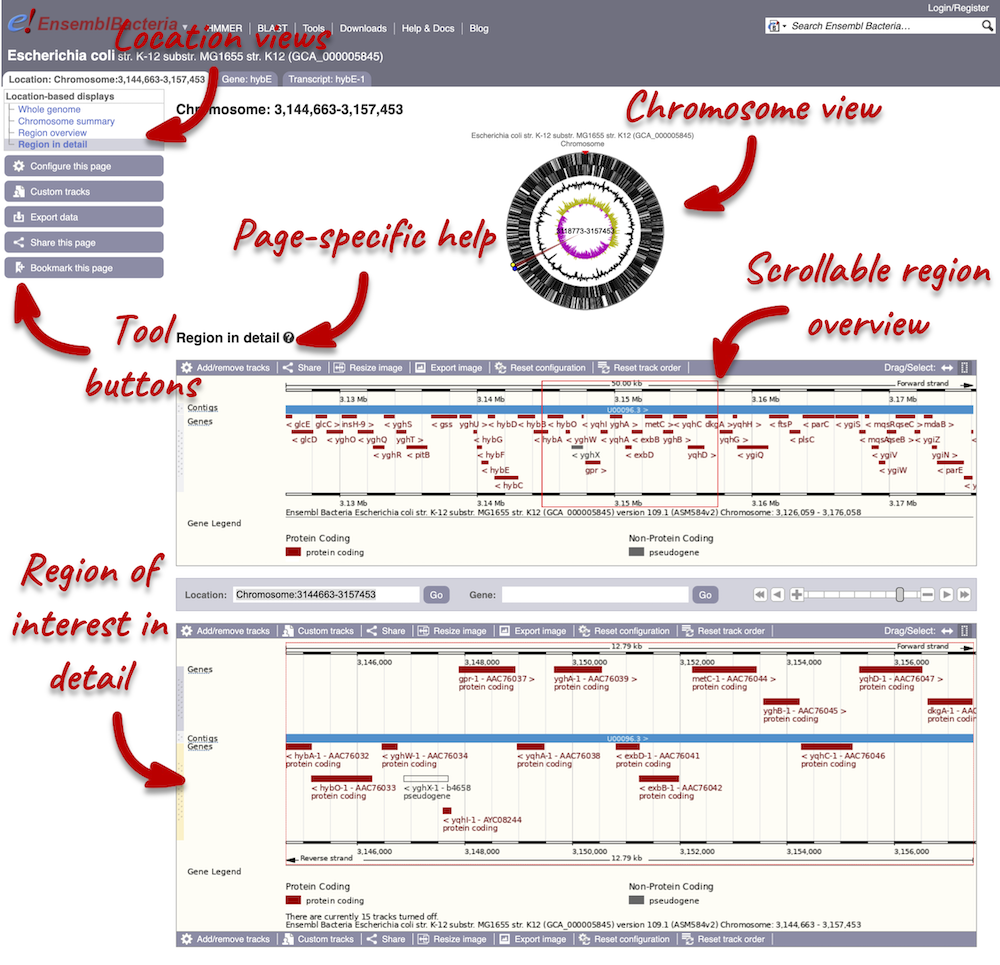
The Region in detail page is made up of three images, let’s look at each one on detail.
The first image shows the chromosome:
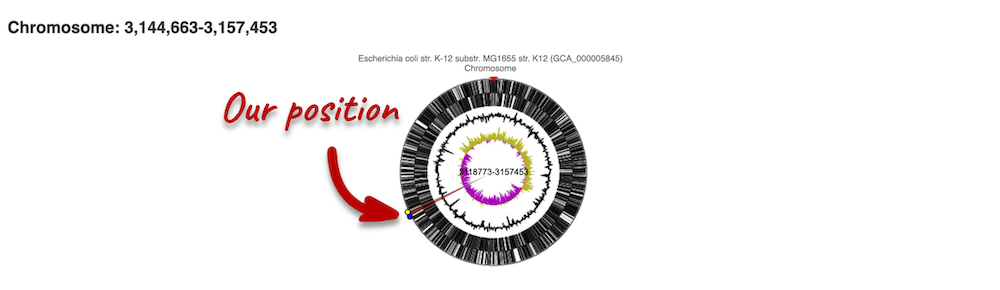
You can jump to a different region by clicking and dragging the yellow and blue handles.
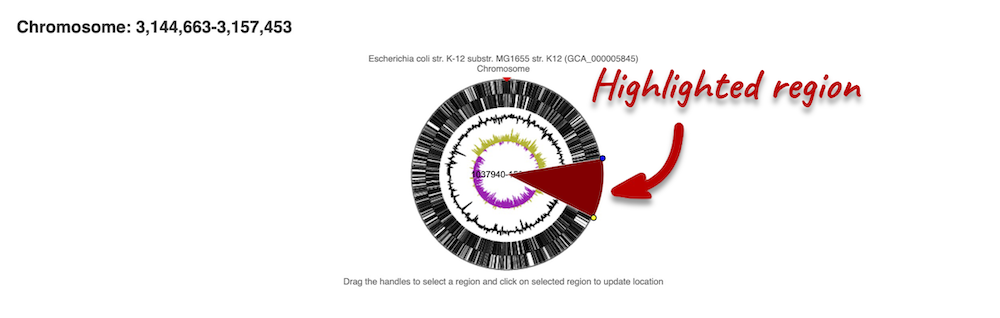
If you want to move to your highlighted region, you click on the region shaded in red.
The second image shows a 50 kb region (the size varies per genome and depends on the gene size and density; you can find a scale at the top of the view) around our selected region. This view allows you to scroll back and forth along the chromosome.
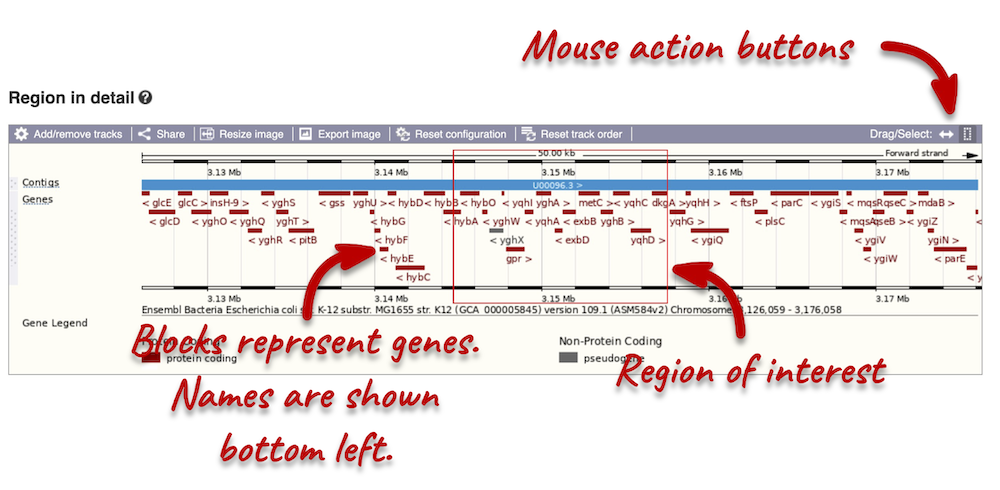
Click and drag your mouse to highlight a region. A pop-up window will appear with options to jump to or centre on the highlighted region.
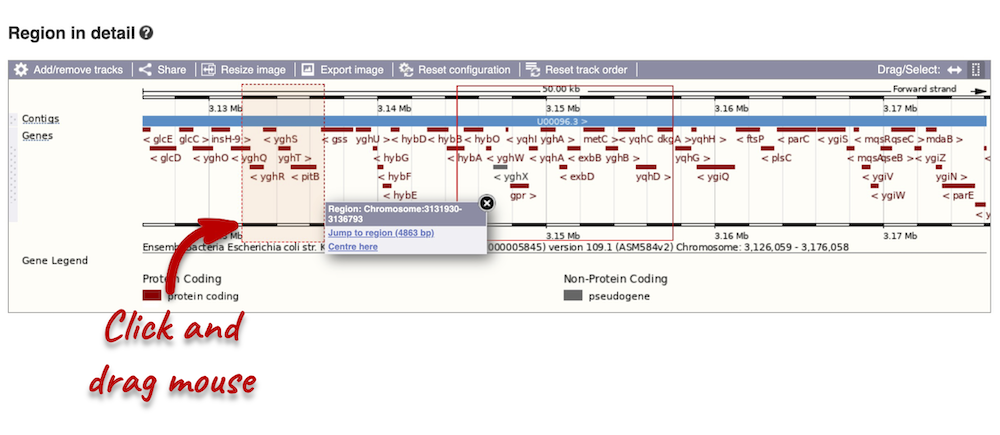
Click on the X to close the pop-up menu.
Click on the Drag/Select button  to change the action of your mouse click. Now you can scroll along the chromosome by clicking and dragging within the image. As you do this you’ll see the image below grey out and update to your scrolled region. To go back to go back to where you started, you can click the Back button of your browser.
to change the action of your mouse click. Now you can scroll along the chromosome by clicking and dragging within the image. As you do this you’ll see the image below grey out and update to your scrolled region. To go back to go back to where you started, you can click the Back button of your browser.
The third image is a detailed, configurable view of the region.
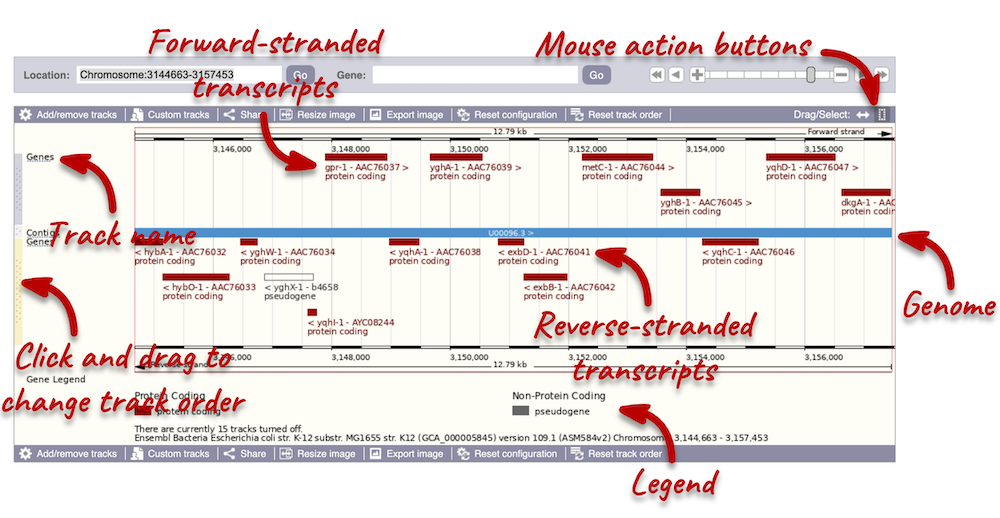
Click on the Drag/Select option at the top or bottom right to switch mouse action. On Drag, you can click and drag left or right to move along the genome, the page will reload when you drop the mouse button. On Select you can drag out a box to highlight or zoom in on a region of interest.
We can edit what we see on this page by clicking on the blue Configure this page menu at the left.

This will open a menu that allows you to change the image.
You can enable tracks in different styles; more details are in the FAQs. 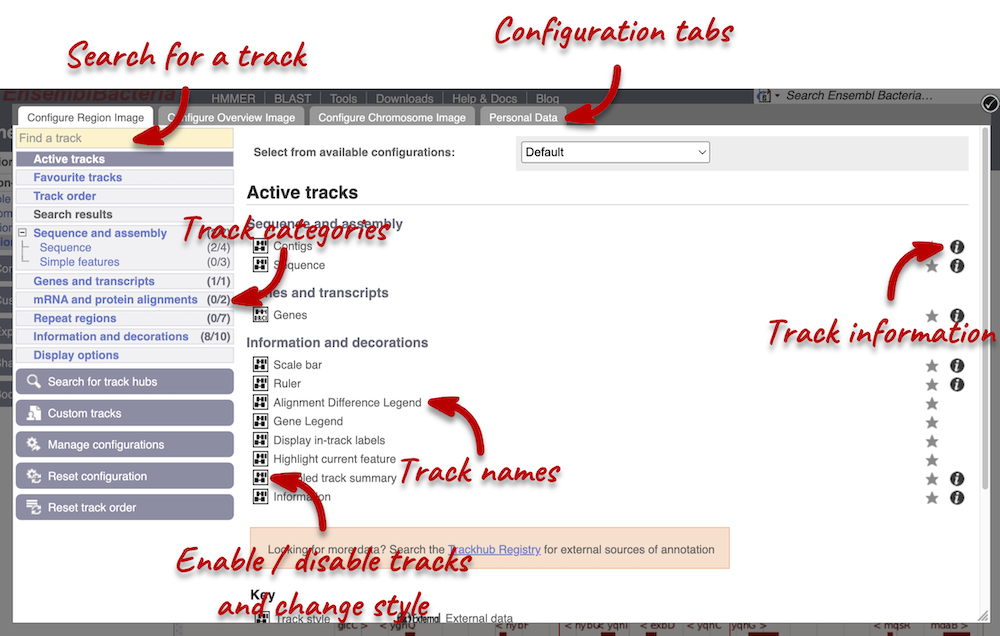
Let’s add the following tracks to our view:
- Start/stop codons
- All repeats
Now click on the check icon in the top left-hand corner to save and close the menu. Alternatively, click anywhere outside of the menu. We can now see the tracks in the image.
We can also change the way the tracks appear by clicking on the track name then hovering over the cog wheel to open its menu. We can move tracks around by clicking and dragging on the bar to the left of the track name.
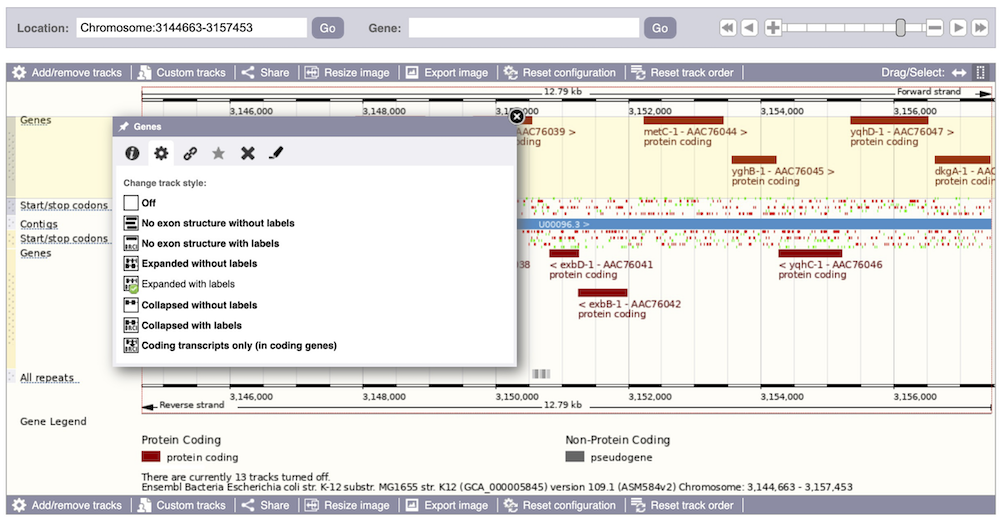
Now that you’ve got the view how you want it, you might like to show something you’ve found to a colleague or collaborator. Click on the Share this page button to generate a URL with your set configurations. Email the link to someone else, so that they can see the same view as you, including all the tracks you’ve added. These links contain the Ensembl release number, so if a new release or even assembly comes out, your link will just take you to the archive site for the release it was made on.
To return this to the default view, go to Configure this page and select Reset configuration at the bottom of the menu.
Demo: Viewing genes and transcripts
You can find out lots of information about Ensembl genes and transcripts using the browser. If you’re already looking at a Region in detail view, you can click on any transcript and a pop-up menu will appear, allowing you to jump directly to that gene or transcript.
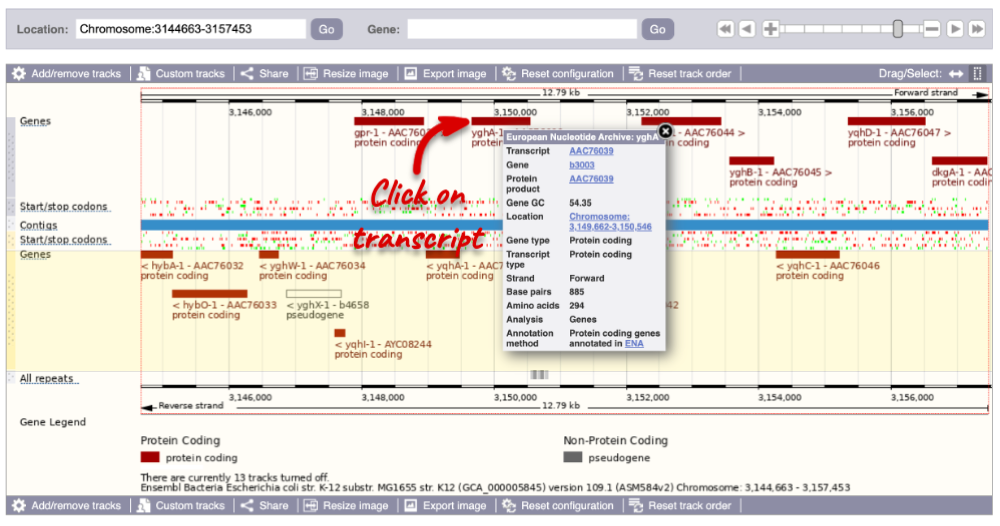
Alternatively, you can find a gene by searching for it. You can search for gene names, identifiers, or functions that might be associated with the genes.
We’re going to look at the lacZ gene Escherichia coli str. K-12 substr. MG1655 (GCA_000005845). From bacteria.ensembl.org, search for the Escherichia coli_ str. K-12 substr. MG1655 (GCA_000005845) genome. Type lacZ into the species-specific search bar and click the Go button.
The gene tab
Click on the gene ID b0344 from the search hits. The Gene tab should open:
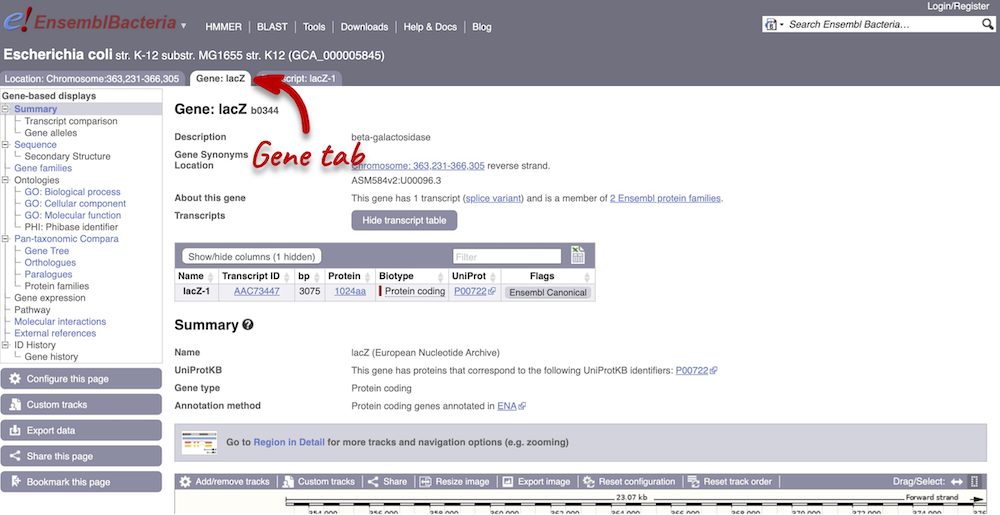
This page summarises the gene, including its location, name and equivalents in other databases. At the bottom of the page, a graphic shows a Region in detail view with the transcripts. We can also see the overlapping and neighbouring genes.
There are different tabs for different types of features, such as genes and transcripts. These appear side-by-side underneath the species name at the top of the page, allowing you to jump back and forth between features of interest. Each tab has its own navigation column down the left hand-side of the page, listing all the things you can see for this feature.
Gene sequence
Let’s walk through the menu for the Gene tab. Click Sequence in the left-hand panel to view the genomic sequence of the gene.
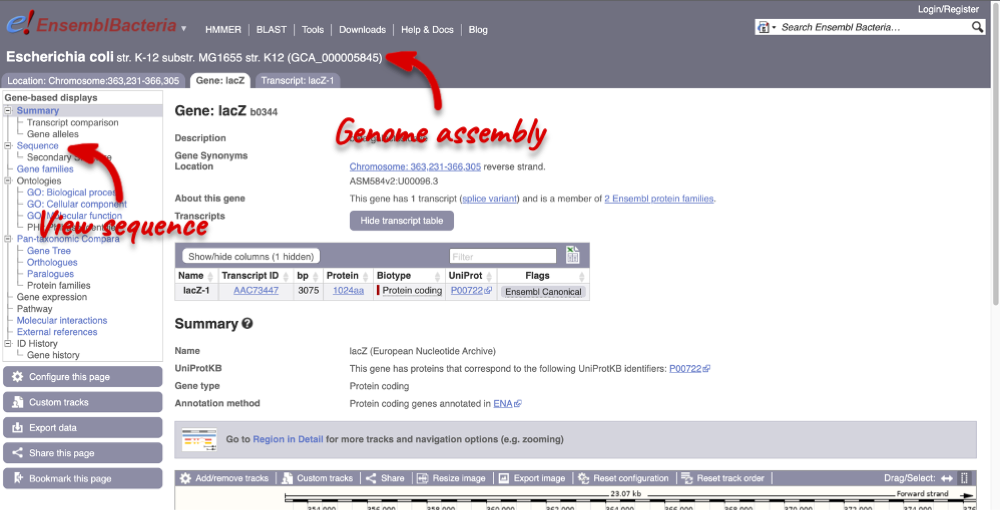
The sequence is shown in FASTA format. The FASTA header contains the genome assembly, chromosome, coordinates and strand (1 or -1). This gene is on the positive strand.

Exons are highlighted within the genomic sequence: the exon of our gene of interest and any neighbouring or overlapping genes. By default, 600 bases are shown up and downstream of the gene. We can make changes to how this sequence appears with the Configure this page button found at the left. This allows us to change the flanking regions, add line numbering and more. Click on it now.
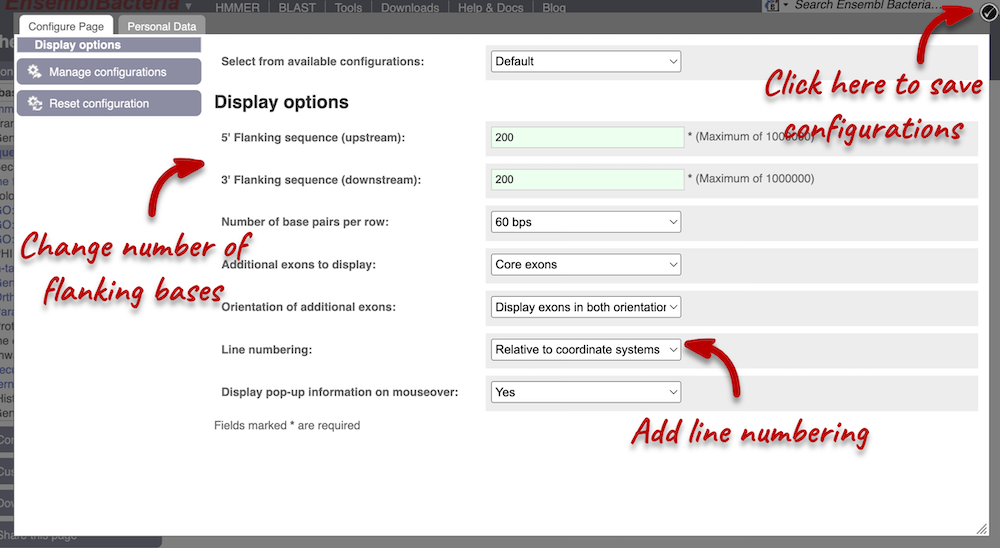
We have changed our Flanking sequences to 200 and added Line numbering relative to the coordinate system. Save your setting by clicking the check icon at the top right-hand corner.
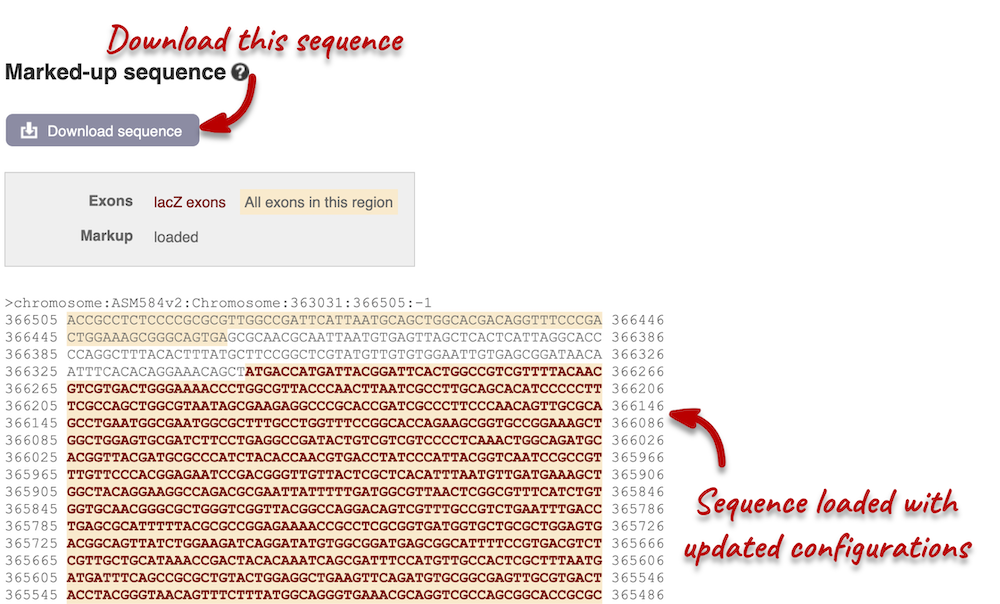
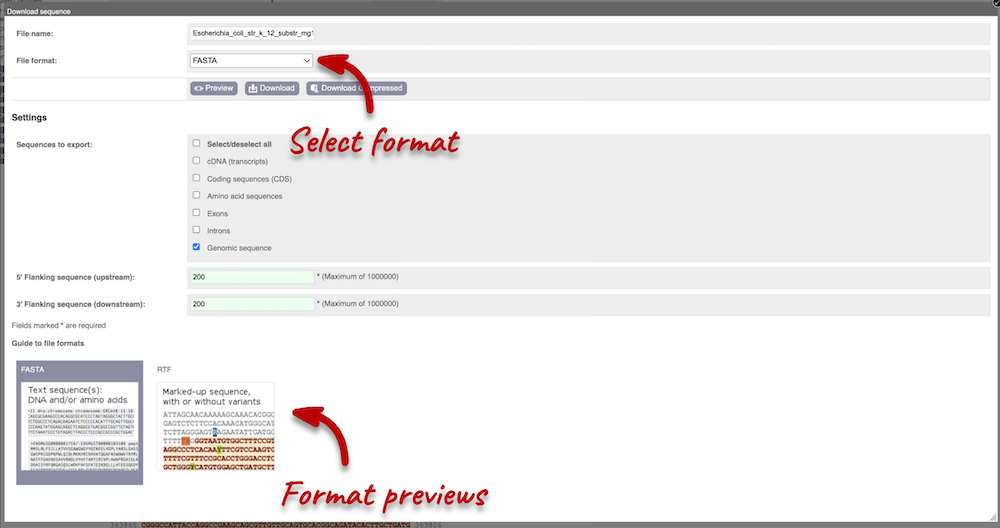
You can download this sequence by clicking in the Download sequence button above the sequence. This will open a dialogue box that allows you to pick between plain FASTA sequence, or sequence in rich-text format (RTF), which includes all the coloured annotations and can be opened in a word processor. If you want run a sequence analysis tool, download as FASTA sequence, whereas if you want to analyse the sequence visually, RTF is best for this. This button is available for all sequence views.
Gene function
To find out the protein function, have a look at gene ontology (GO) terms from the Gene Ontology consortium. There are three pages of GO terms, representing the three divisions: GO: Biological process (what the protein does)
GO: Cellular component (where the protein is)
GO: Molecular function (how it does it)
Click on GO: Biological process to see an example of the GO pages.
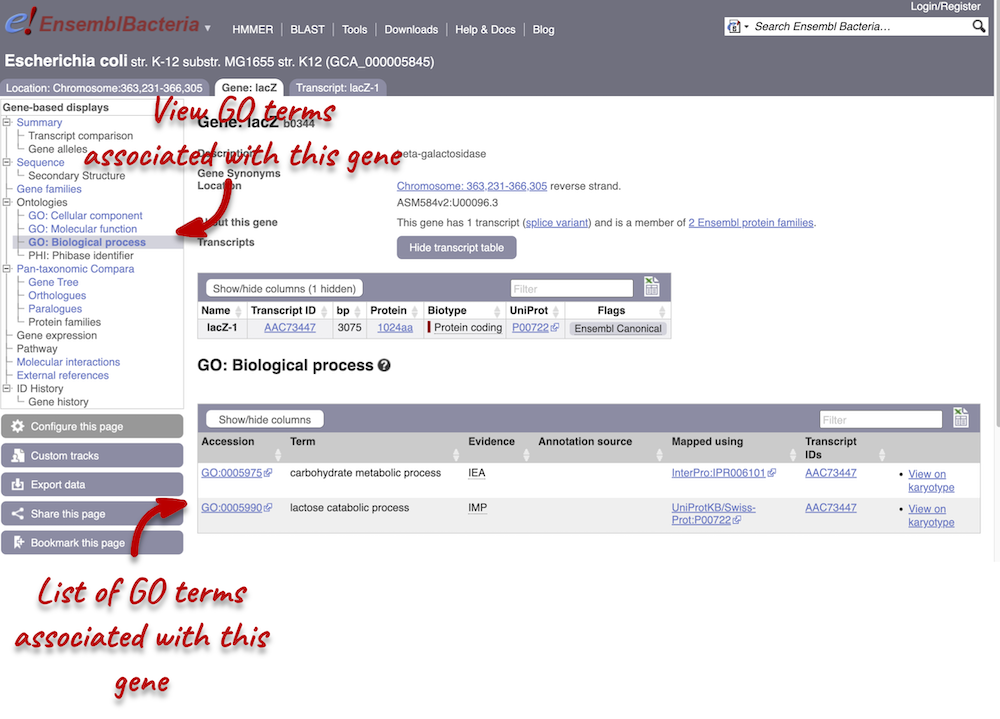
Here you can see the functions that have been associated with the gene. There are three-letter codes that indicate how the association was made, as well as links to the specific transcript they are linked to.
Gene information in external databases
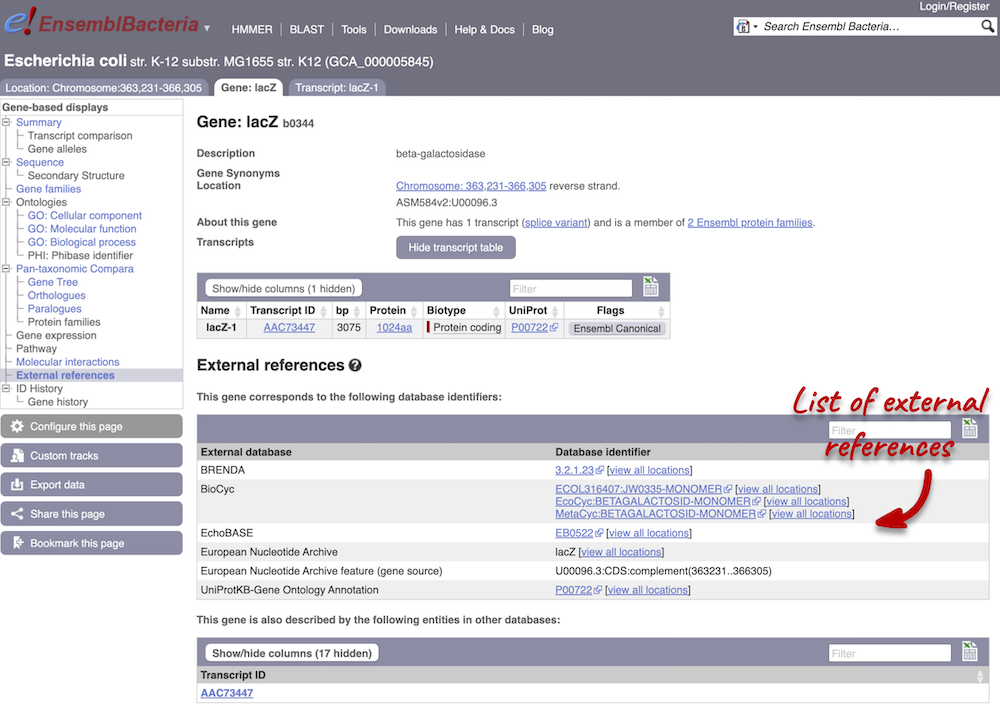
We also have links out to other databases which have information about our genes and may focus on other topics that we don’t cover, like the European Nucleotide Archive ENA) or the UniProt knowledge base UniProtKB. Go up the left-hand menu to External references:
The transcript tab
We’re now going to explore the transcript of lacZ. Click on Show transcript table underneath the gene summary at the top of the page.

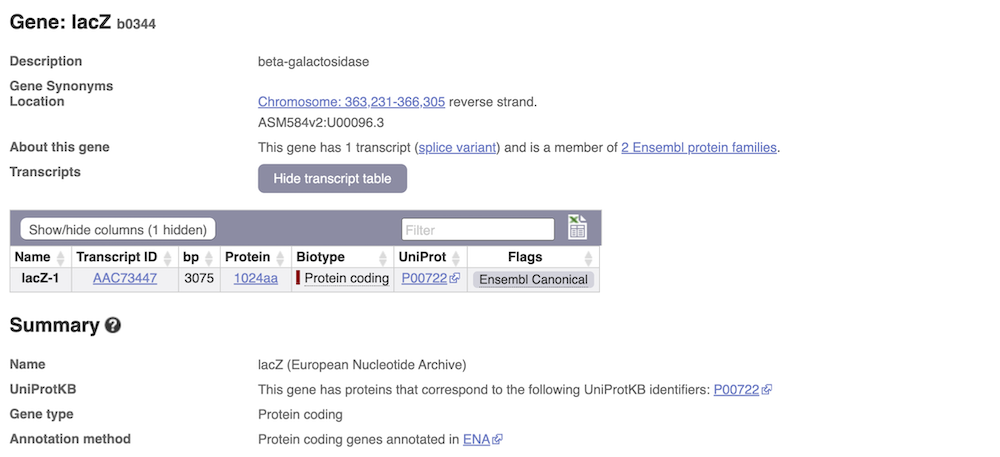
Here we can see a list of all the transcripts of lacZ with their identifiers, lengths and biotypes. The lacZ gene only has one transcript. Click on the transcript ID AAC73447.
You are now in the Transcript tab for AAC73447. We can still see the gene tab so we can easily jump back. The left hand navigation column provides several options for the transcript AAC73447 - many of these are similar to the options you see in the gene tab, but not all of them. If you can’t find the thing you’re looking for, often the solution is to switch tabs.
Transcript sequences
Click on the Exons link in the left-hand panel. This page is useful as it will give you the length of the coding sequence.
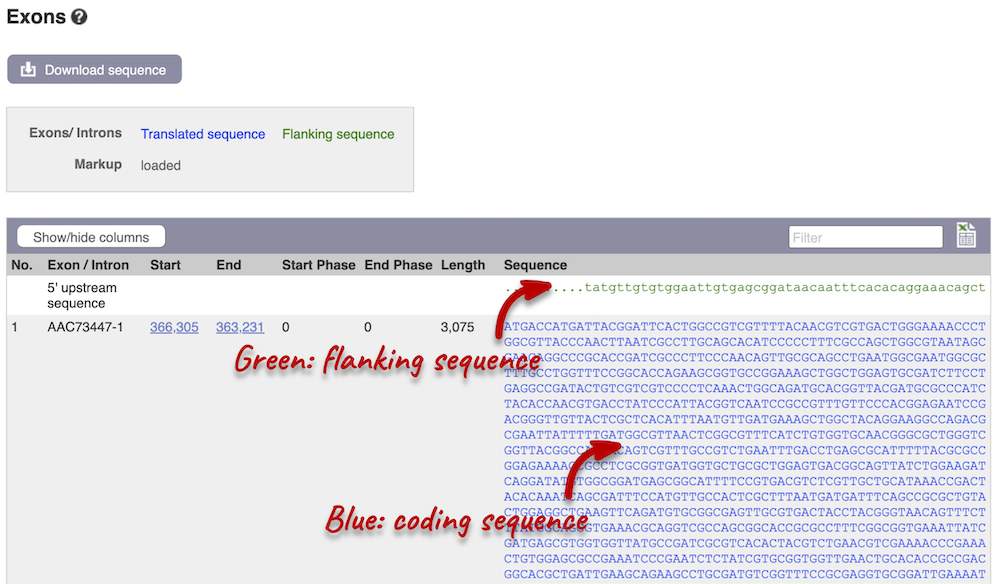
You may want to change the display (for example, to show more flanking sequences). In order to do so, click on Configure this page and change the display options accordingly.
Transcript information in external databases
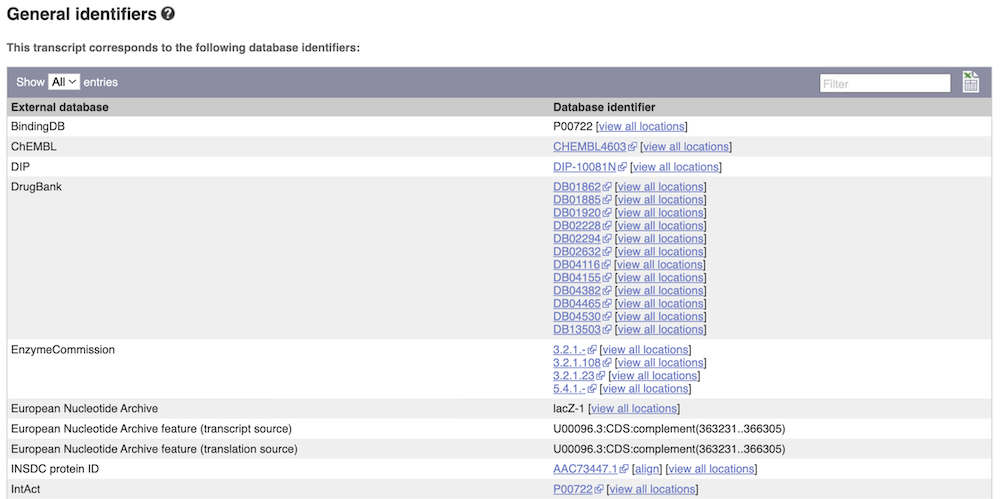
Next, follow the General identifiers link at the left. Just like the External References page in the Gene tab, this page shows links out to other databases such as InterPro, PDB, UniProtKB, and others, this time linked to the transcript or protein product, rather than the gene.
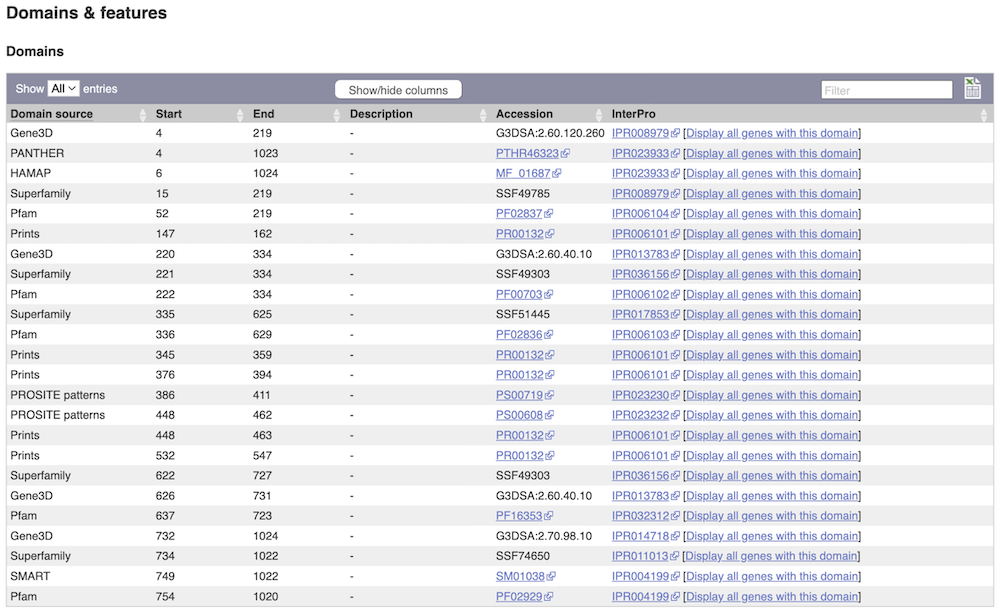
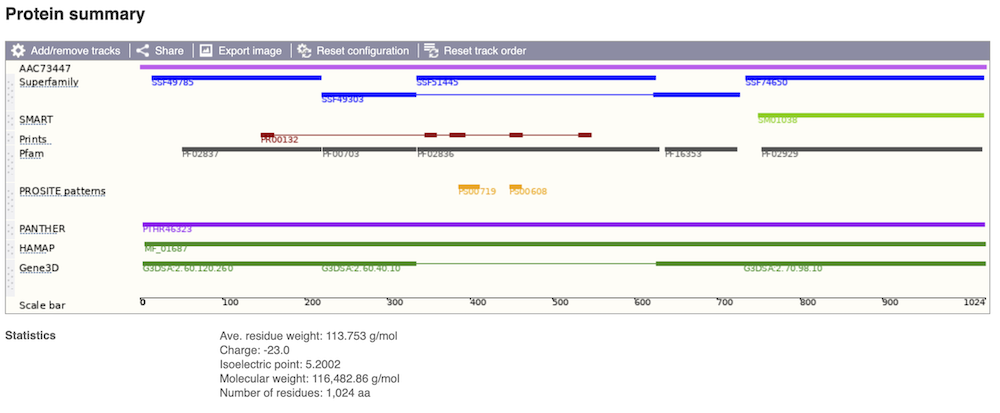
Protein domain information
If you’re interested in protein domains, you could click on Protein summary to view domains from different sources, such as SMART and PROSITE. These are all plotted against the transcript sequence.
Alternatively, you can go to Domains & features to see a table of the same information in a tabular format.