Filter Events by Year
Webinar: Explore Human Pangenome Reference Consortium (HPRC) data in Ensembl
Course Details
- Lead Trainer
- Louisse Paola Mirabueno
- Associate Trainer(s)
- Event Date
- 2024-03-28
- Location
- Virtual
- Description
- The Human Pangenome Reference Consortium (HPRC) aims to construct a comprehensive and representative human pangenome reference to better illustrate the genomic landscape of diverse human populations. In this webinar, we will guide you on how to access HPRC data via the Ensembl genome browser. We will explore the available gene annotation and genetic variation data of the sequence assemblies produced by the project.
Demos and exercises
HPRC data in Ensembl
Demonstration: accessing HPRC data in Ensembl Rapid Release
Available HPRC genome assemblies in Ensembl
All available Human Pangenome Reference Consortium (HPRC) genomes can be found on the Ensembl Projects page. Currently, gene annotation, genetic variation and homology data of 96 haplotypes as well as the T2T-CHM13v2.0 assembly are available through the Ensembl Rapid Release browser and the Ensembl FTP site.
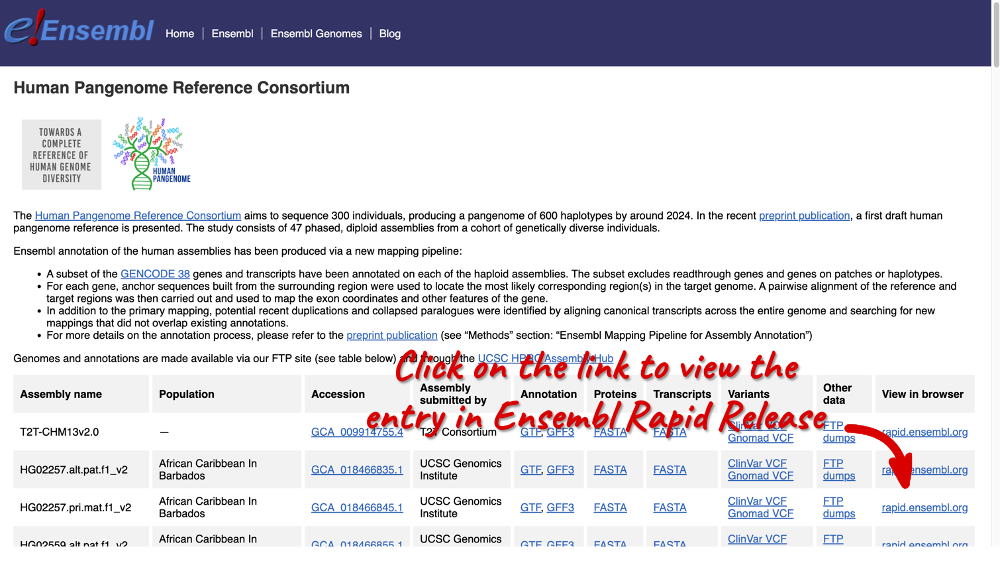
You can click through to the species information page in Ensembl Rapid Release by selecting a specific assembly from the table and clicking on the rapid.ensembl.org link.
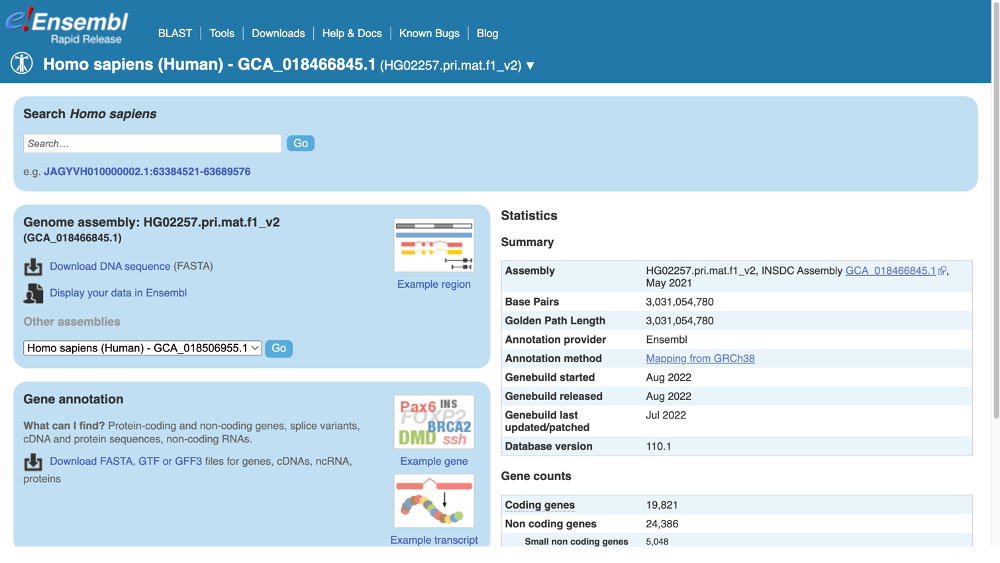
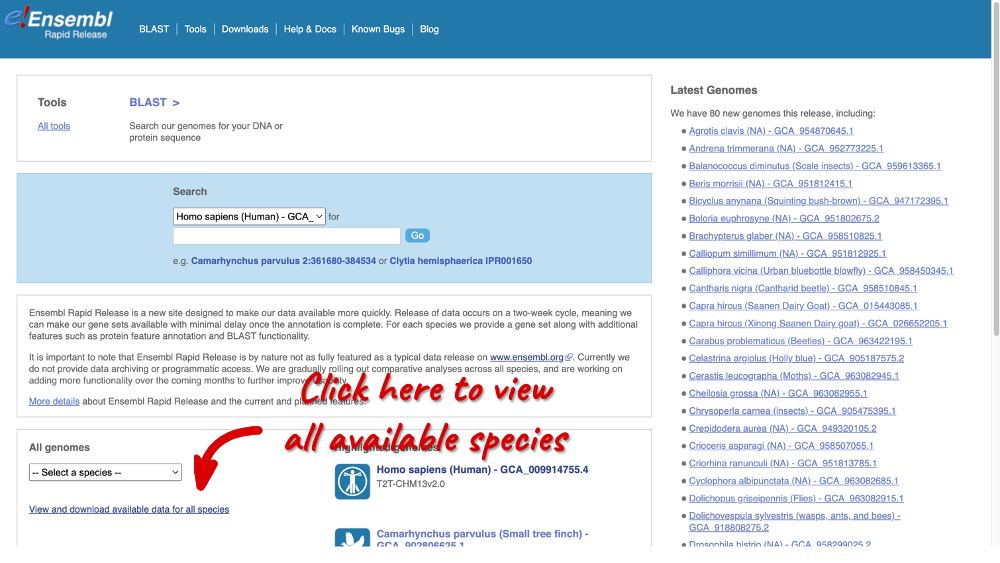
Alternatively, you can open the Ensembl Rapid Release website and click on View and downloadd available data for all species.
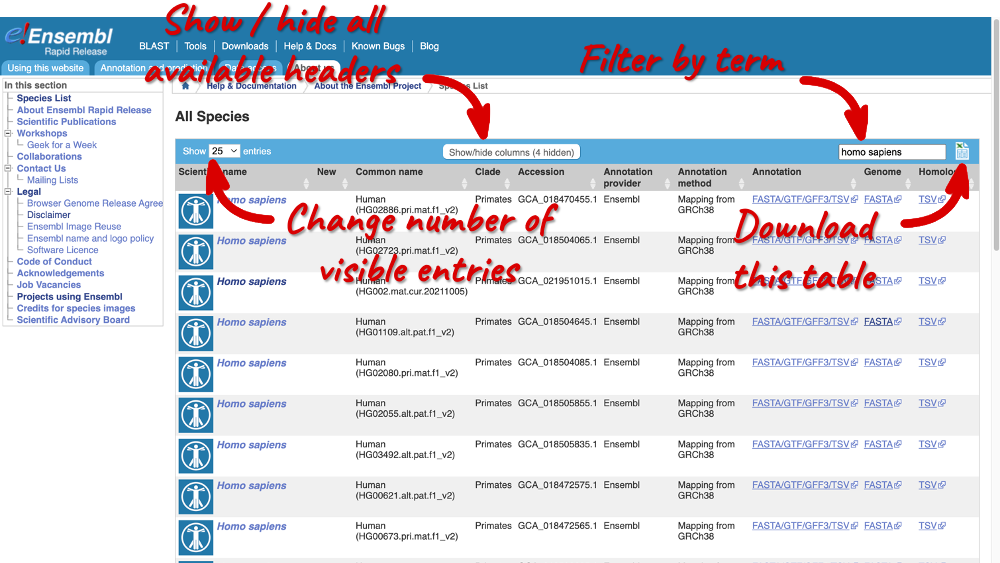
Filter the table by entering Homo sapiens in the text box in the top right-hand corner to find all available HPRC assemblies in Ensembl. The table includes genome accession and annotation sources. There are also links out to the FTP site where you can download whole genome flatfiles. There are annotation files with the loci and sequences of the genes, the whole genome sequence and BAM files. The BAM files are the RNA-seq data that was used to annotate the genes.
You can change the number of visible entries by clicking on the drop-down menu, display or hide columns by clicking on Show/hide collumn, or download the table by clicking on the Excel icon.
Click on any FASTA link. In many internet browsers, this will try to open in an FTP client, so you may need to right-click on FASTA to copy the link address and change the ftp at the beginning of the URL to https to open the site on your browser.
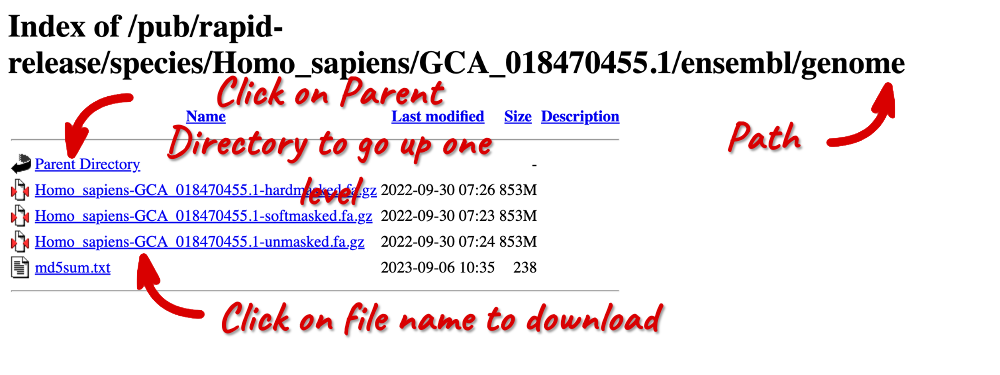
This is the FTP page for the assembly you selected. Here, you can find hard-masked, soft-masked and unmasked FASTA files. You can read more about repeat masking here.
The Gene tab
Go back to the species list, sort the table by Common name and click on Homo sapiens for the maternal haplotype of HG02886 (GCA_018470455.1), a genome of Gambian ancestry.
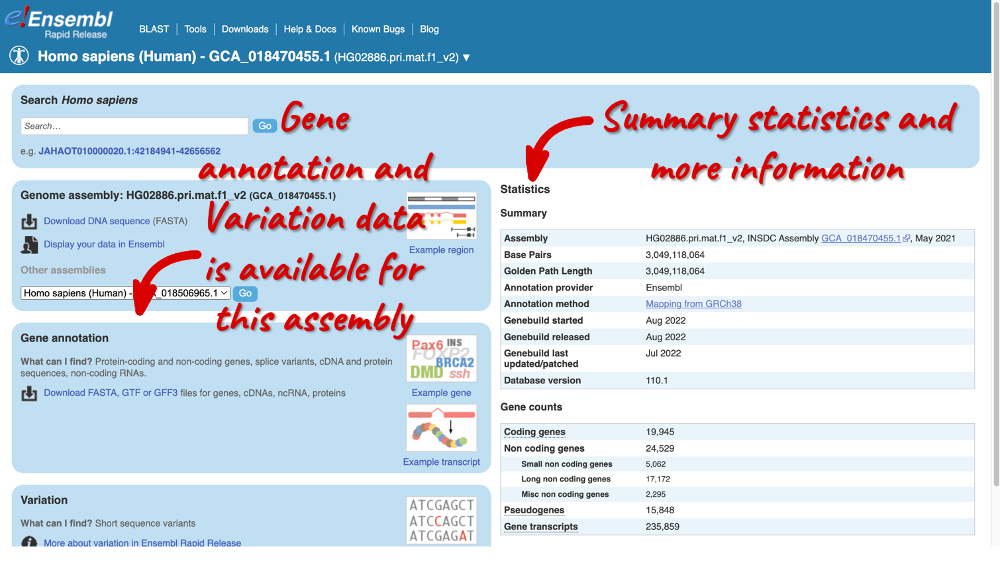
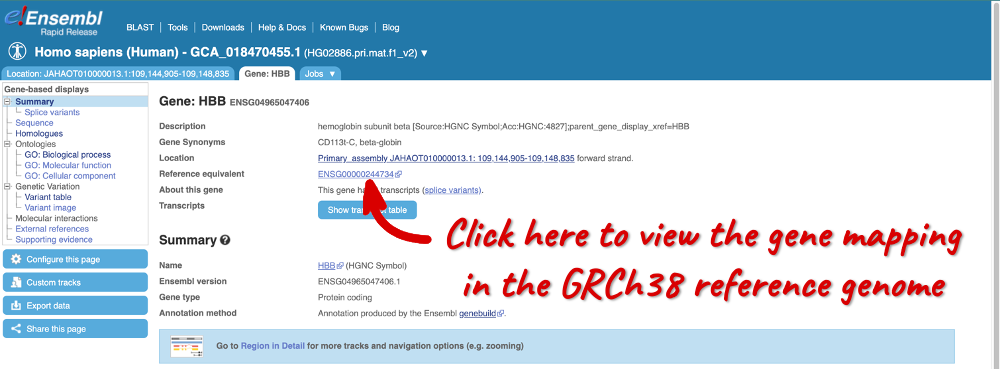
Here you can see links to example data and statistics about the genome. Let’s search for a gene: enter HBB in the search bar (the corresponding Ensembl ID is ENSG04965047406).
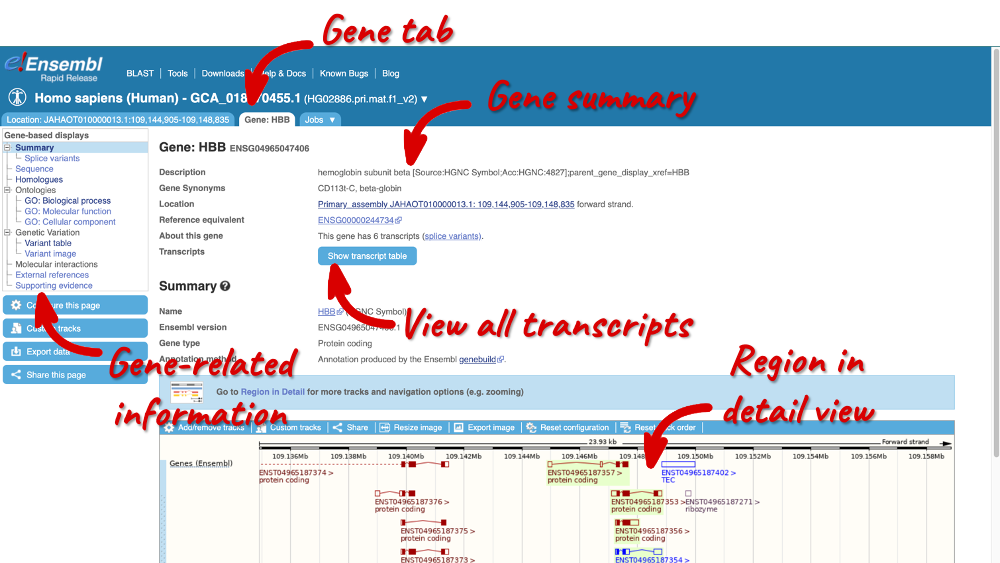
Clicking on the gene ID opens the Gene tab, where you can find Summary information about the gene, a table of its transcripts and a visualisation of the genomic region.
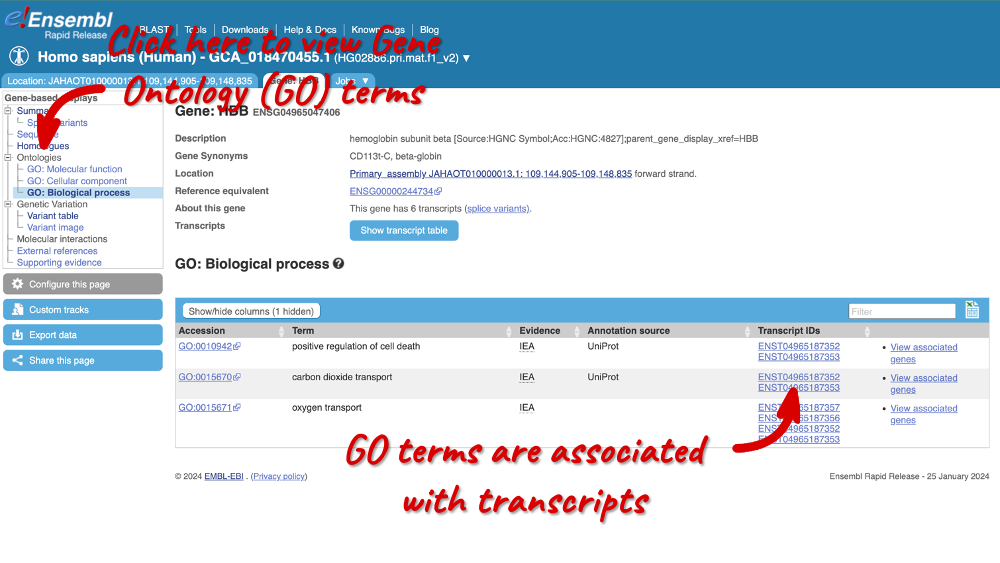
There is a limited amount of annotation attached to the gene. There are GO terms, which were assigned based on protein domains from sequence analysis. Click on any of the GO categories to see the terms.
Reference (GRCh38) equivalent (also available in GTF under projection_parent_gene flag)
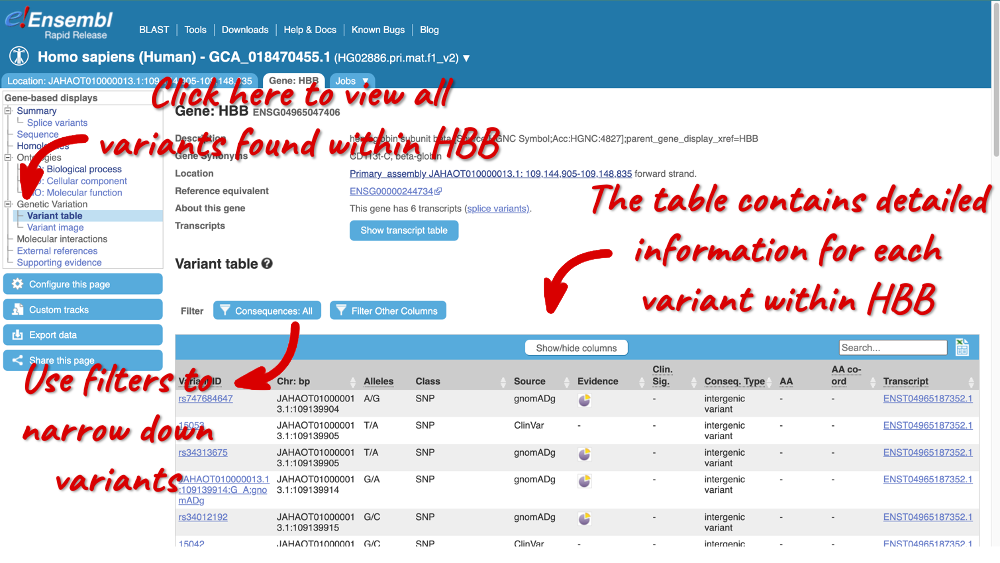
Variant table
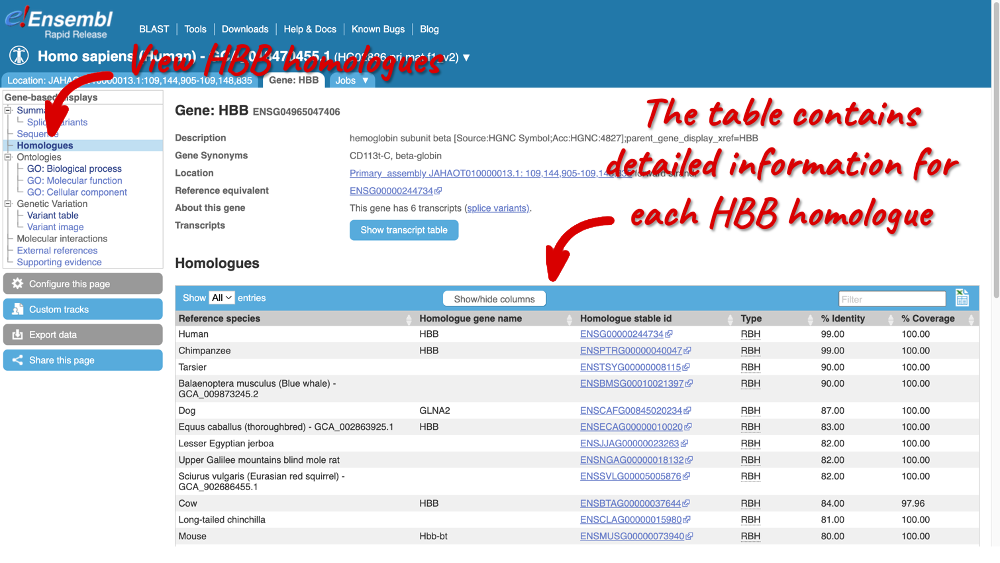
Homologues
The Location tab
Let’s explore the Location tab by clicking on Location: JAHAOT010000013.1: 109,144,905-109,148,835 in the top left-hand corner. The Region in detail view is composed of three resolutions of the genomic region on the right: the top image shows chromosome overview, the middle image shows a region overview displaying flanking regions within 1.00Mb of the HBB gene (highlighted in green) and the Region in detail view showing individual transcripts and more detailed features within the JAHAOT010000013.1: 109,144,905-109,148,835 region.
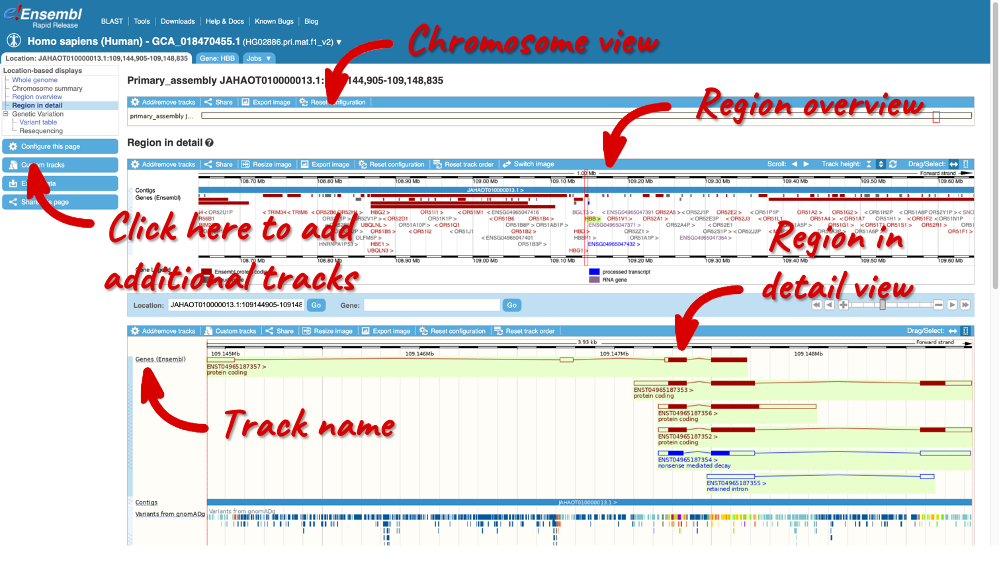
You can visualise various genetic features within this display. Let’s add variant data across this locus by clicking on Configure this page.
This pop-up window menu shows us all the possible tracks you can view. Click on Variation in the left-hand menu. Click on the i on the far right to view a description of each track. Click on the boxes on the let of the track name to add the data individually. Close the pop-up window by clicking on the the Check icon in the top right-hand corner or anywhere outside the pop-up.








