Filter Events by Year
Ensembl Browser Workshop - Fountain University
Course Details
- Lead Trainer
- Ben Moore
- Event Date
- 2023-01-25
- Location
- Virtual: Fountain University, Osogbo, Nigeria
- Description
- Work with the Ensembl Outreach team to get to grips with the Ensembl browser, accessing gene, variation, comparative genomics and regulation data, and mine these data with BioMart.
- Survey
- Ensembl Browser Workshop - Fountain University Feedback Survey
Demos and exercises
Species and genome assemblies
Demo: Introduction to Ensembl
Ensembl
Homepage
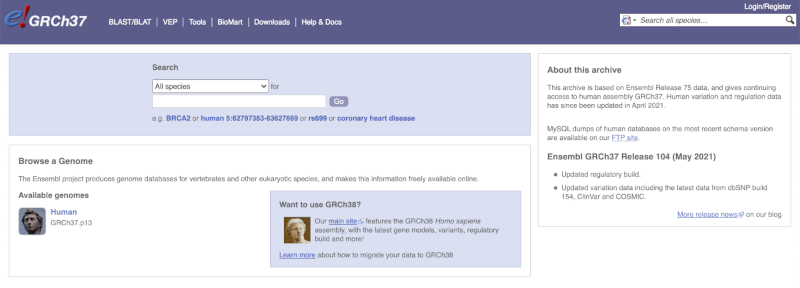
The front page of Ensembl is found at ensembl.org. It contains lots of information and links to help you navigate Ensembl:
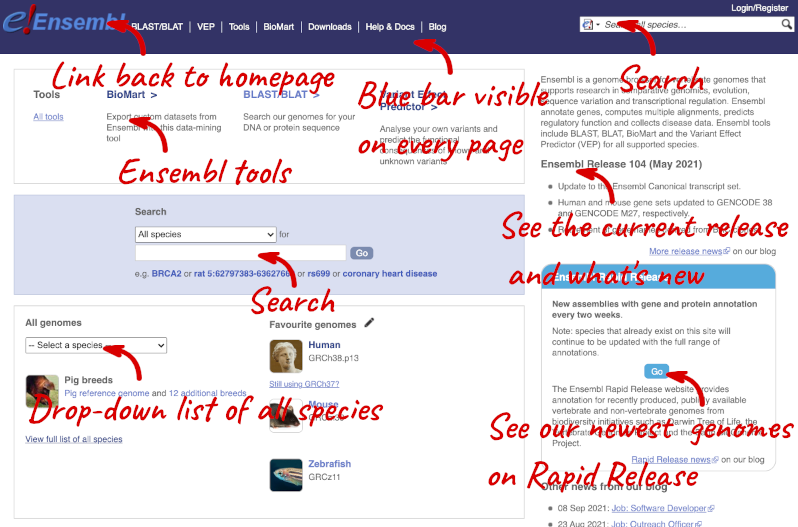
On the right-hand panel you can see the current release number and what has come out in this release. To access old releases, scroll to the bottom of the page and click on View in archive site in the right-hand corner.
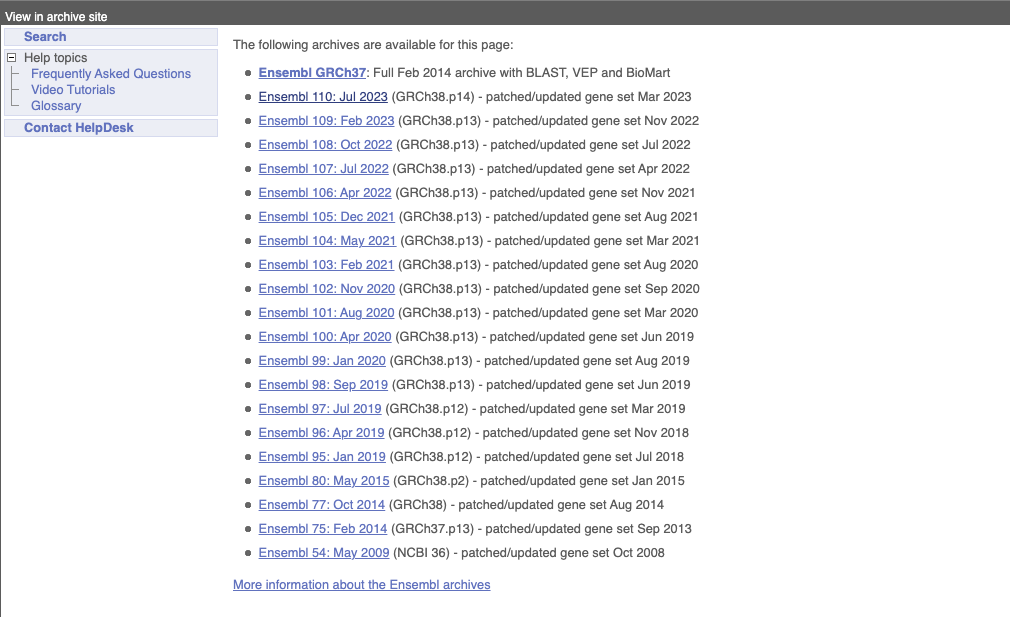
Click on the links to go to the archives. Alternatively, you can jump quickly to the correct release by adding e plus the release number in the URL. For example e98.ensembl.org jumps to Ensembl release 98.
Available species
Scroll back up to the top of the homepage. You can view all available species by clicking the View full list of all species link underneath the coloured search block.
You can search for your species of interest (either the common or scientific name) using the search bar at the top right-hand corner of the table. Click on the common name of your species of interest to go to the species information page. We’ll click on Human.
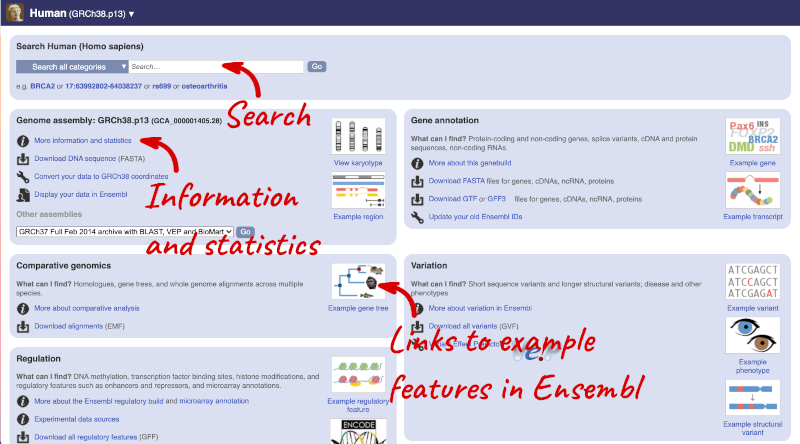
Species information
Here you can see links to example features and to download flatfiles. To find out more about the genome assembly and genebuild, click on More information and statistics under the Genome assembly section.
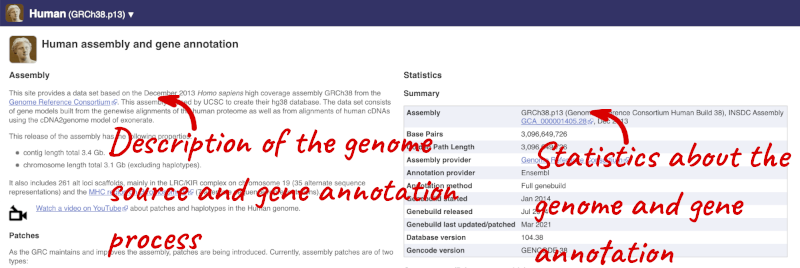
Here you’ll find a detailed description of how to the genome was produced and links to the original source. You will also see details of how the genes were annotated.
The current genome assembly for human is GRCh38. If you want to see the previous assembly, GRCh37, visit our dedicated site grch37.ensembl.org.
Ensembl Genomes
Homepage
Let’s take a look at the Ensembl Genomes homepage at ensemblgenomes.org.
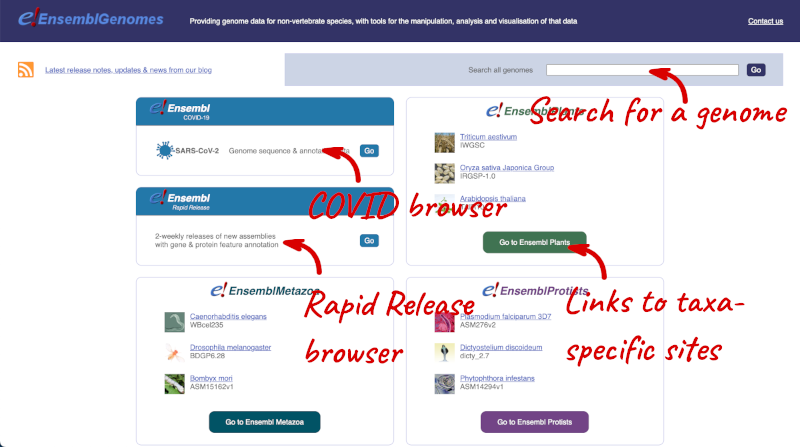
Click on the different taxa to see their homepages. Each one has a different colour-coding, but they are all structured in a similar format to the Ensembl main site.
You can navigate most of the taxa in the same way as you would with Ensembl.
Ensembl Bacteria
Ensembl Bacteria has a large number of genomes and has a slightly different method to the other Ensembl sites. Let’s look at it in more detail.
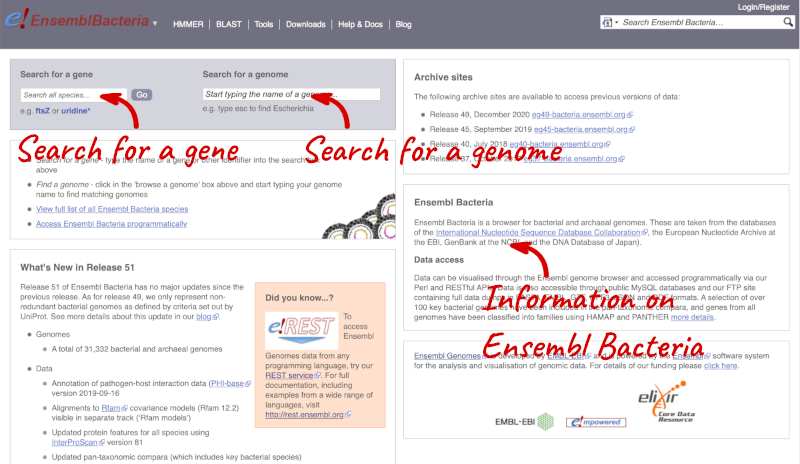
There’s no drop-down species list for bacteria as it would be hard to navigate with the number of species. You can click the View full list of all Ensembl Bacteria species link underneath the coloured search block. Search for your species of interest using the filter in the top right-hand corner of the table.
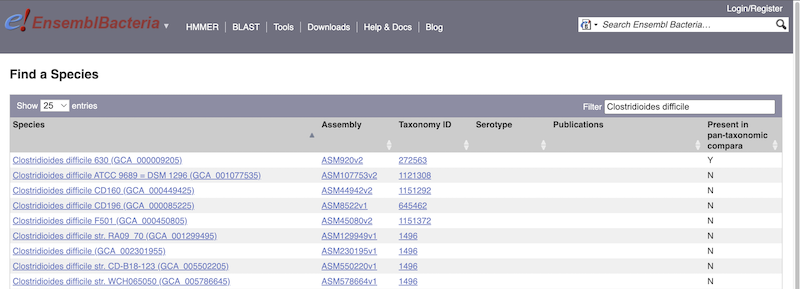
Alternatively, you can find a species by typing the species name into the Search for a genome search box at the top of the page. A drop-down list will appear with any species matching the name you entered.
For example, to find a sub-strain of Clostridioides difficile start typing in the species name. Due to the auto-complete, you’ll see useful results as soon as you get to Clostridio.
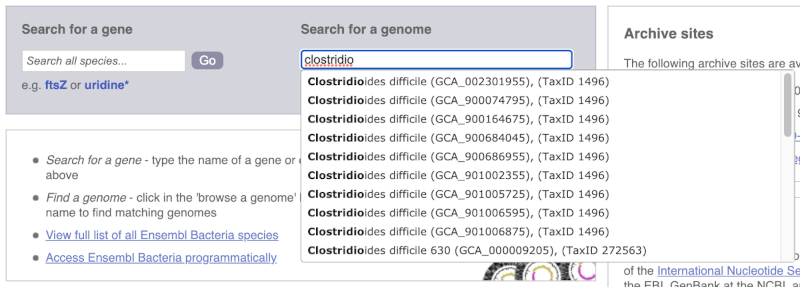
The drop down contains various strains of C. difficile. Let’s choose C. difficile 630. This will take us to another species information page, where we can explore various features.
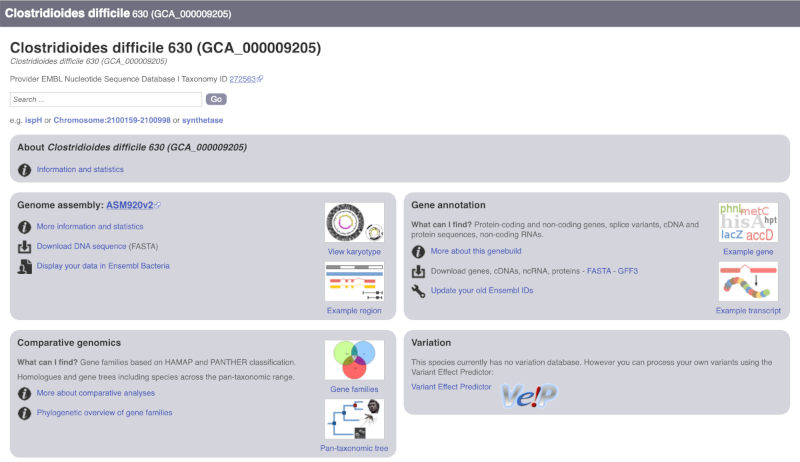
Unlike the Homo sapiens species information page, there is no prose description of the genome or gene annotation, as these pages were generated automatically.
Ensembl Rapid Release
Our newest genomes, such as those coming from the Darwin Tree of Life, are available rapid.ensembl.org with limited annotation.
Panda species
Go to Ensembl and find the following information:
-
What is the name of the genome assembly for Panda?
-
How long is the Panda genome (in bp)? How many coding genes have been annotated?
-
Select Giant panda from the drop down species list, or click on View full list of all Ensembl species, then choose Giant panda from the list.
The assembly is ASM200744v2 or GCA_002007445.2. -
Click on More information and statistics. Statistics are shown in the tables on the left.
The length of the genome is 2,444,060,653 bp.
There are 20,857 coding genes.
Available zebrafish assemblies
What previous assemblies are available for zebrafish?
Click on Zebrafish on the front page of Ensembl to go to the species homepage. Under Other assemblies three previous assembly names and the releases you can find them in are listed.
Assembly GRCz10 is available in the archived release 80, Zv9 in 77 and Zv8 in 54.
Solanum genus
Go to Ensembl Plants and answer the following questions:
-
How many genomes of the genus Solanum are there in Ensembl Plants?
-
When was the current Solanum lycopersicum genome assembly last revised?
- On the homepage, click on View full list of all Ensembl Plants species underneath the coloured search block. Type Solanum into the filter box in the top left-hand corner of the table.
There are three Solanum genomes: Solanum lycopersicum (tomato), and Solanum tuberosum RH89-039-16 and Solanum tuberosum (both potato).
- Click on S. lycopersicum, then on More information and statistics.
The genome was revised in April 2018.
Mosquito species
-
Go to Ensembl Metazoa. How many genomes relating to the genus Anopheles are there in Ensembl Metazoa?
-
When was the current Anopheles gambiae genome assembly last revised?
- Go to metazoa.ensembl.org. Open the drop-down list or click on View full list of all Ensembl Metazoa species. In a latin binomial species name, the first word represents the genus. Type Anopheles into the filter box in the top left to find all genomes with this word in the binomial.
There are 22 Anopheles genomes (some species are represented by more than one genome).
- Click on Anopheles gambiae (African malaria mosquito, PEST), and then on More information and statistics.
The assembly hosted is AgamP4 (INSDC Assembly GCA_000005575.1) which was revised in Feb 2006.
Finding a genome in Ensembl Bacteria
Mycobacterium tuberculosis H37Ra str. ATCC25177 is a clinical strain.
Go to Ensembl Bacteria and find the species M. tuberculosis H37Ra str. ATCC25177. How many coding genes does it have?
In the Ensesmbl Bacteria homepage, start to type H37Ra into the Search for a genome search box (you can find this in the coloured block at the top of the homepage). It will auto-complete, allowing you to select M. tuberculosis H37Ra str. ATCC25177 from the drop-down list. Click on More information and statistics.
M. tuberculosis H37Ra str. ATCC25177 has 4,080 coding and 47 non-coding genes.
Exploring genomic regions
Start at the Ensembl front page, ensembl.org. You can search for a region by typing it into a search box, but you have to specify the species.
To bypass the text search, you need to input your region coordinates in the correct format, which is chromosome, colon, start coordinate, dash, end coordinate, with no spaces for example: human 4:122868000-122946000. Type (or copy and paste) these coordinates into either search box.
Press Enter or click Go to jump directly to the Region in detail Page.
Click on the  button to view page-specific help. The help pages provide text, labelled images and, in some cases, help videos to describe what you can see on the page and how to interact with it.
button to view page-specific help. The help pages provide text, labelled images and, in some cases, help videos to describe what you can see on the page and how to interact with it.
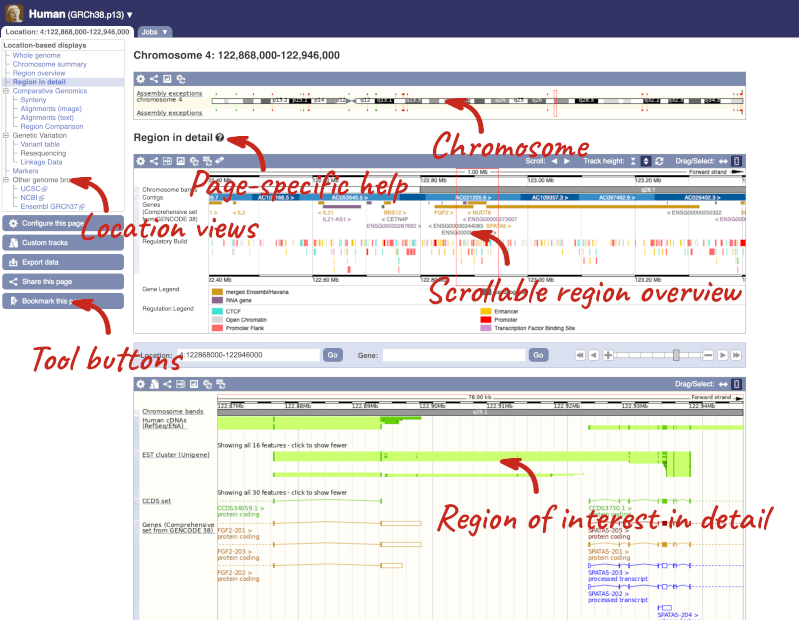
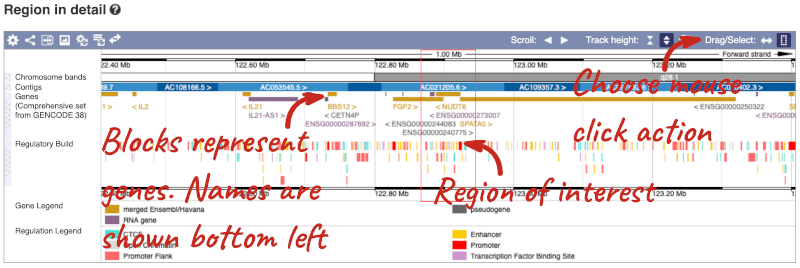
The Region in detail page is made up of three images, let’s look at each one in detail.
The first image shows the chromosome:
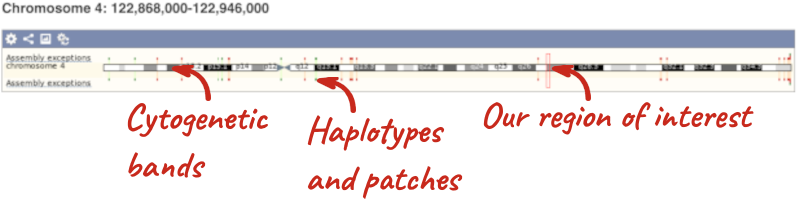
The region we’re looking at is highlighted on the chromosome. You can jump to a different region by dragging out a box in this image. Drag out a box on the chromosome, a pop-up menu will appear.
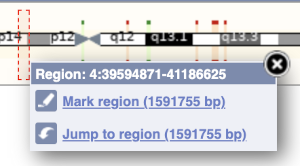
If you wanted to move to the region, you could click on Jump to region (### bp). If you wanted to highlight it, click on Mark region (###bp). For now, we’ll close the pop-up by clicking on the X on the corner.
The second image shows a 1Mb region around our selected region. This is always 1Mb in human, but the fixed size of this view varies between species. This view allows you to scroll back and forth along the chromosome.
You can also drag out and jump to or mark a region.
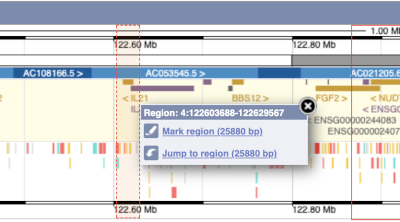
Click on the X to close the pop-up menu.
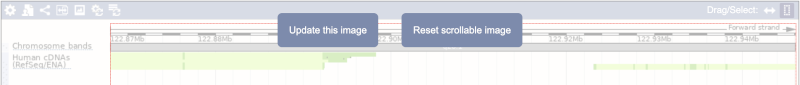
Click on the Drag/Select button  to change the action of your mouse click. Now you can scroll along the chromosome by clicking and dragging within the image. As you do this you’ll see the image below grey out and two blue buttons appear. Clicking on Update this image would jump the lower image to the region central to the scrollable image. We want to go back to where we started, so we’ll click on Reset scrollable image.
to change the action of your mouse click. Now you can scroll along the chromosome by clicking and dragging within the image. As you do this you’ll see the image below grey out and two blue buttons appear. Clicking on Update this image would jump the lower image to the region central to the scrollable image. We want to go back to where we started, so we’ll click on Reset scrollable image.
The third image is a detailed, configurable view of the region.
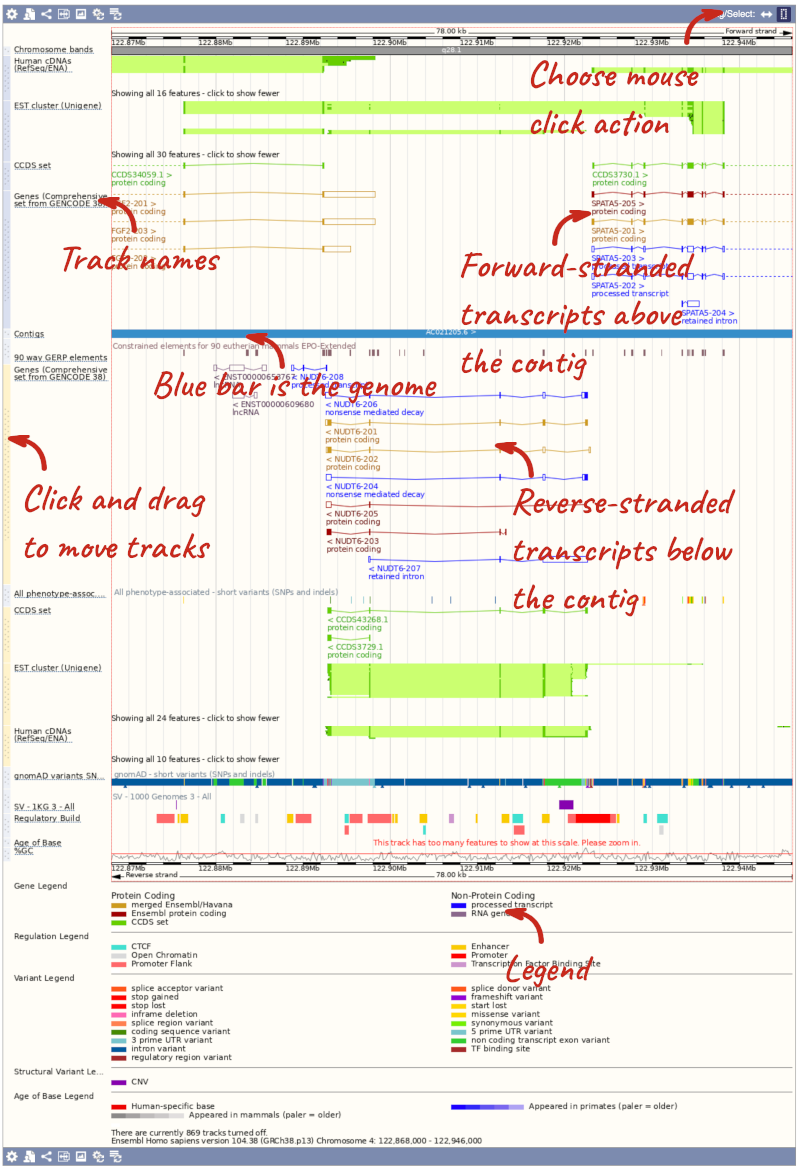
Here you can see various tracks, which is what we call a data type that you can plot against the genome. Some tracks, such as the transcripts, can be on the forward or reverse strand. Forward stranded features are shown above the blue contig track that runs across the middle of the image, with reverse stranded features below the contig. Other tracks, such as variants, regulatory features or conserved regions, refer to both strands of the genome, and these are shown by default at the very top or very bottom of the view.
You can use click and drag to either navigate around the region or highlight regions of interest, Click on the Drag/Select option at the top or bottom right to switch mouse action. On Drag, you can click and drag left or right to move along the genome, the page will reload when you drop the mouse button. On Select you can drag out a box to highlight or zoom in on a region of interest.
With the tool set to Select, drag out a box around an exon and choose Mark region.
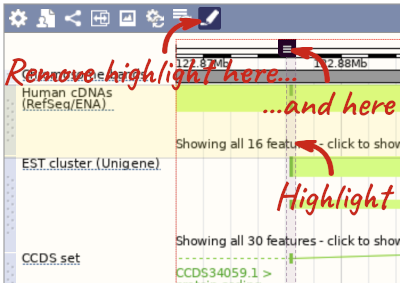
The highlight will remain in place if you zoom in and out or move around the region. This allows you to keep track of regions or features of interest.
We can edit what we see on this page by clicking on the blue Configure this page menu at the left.

This will open a menu that allows you to change the image.
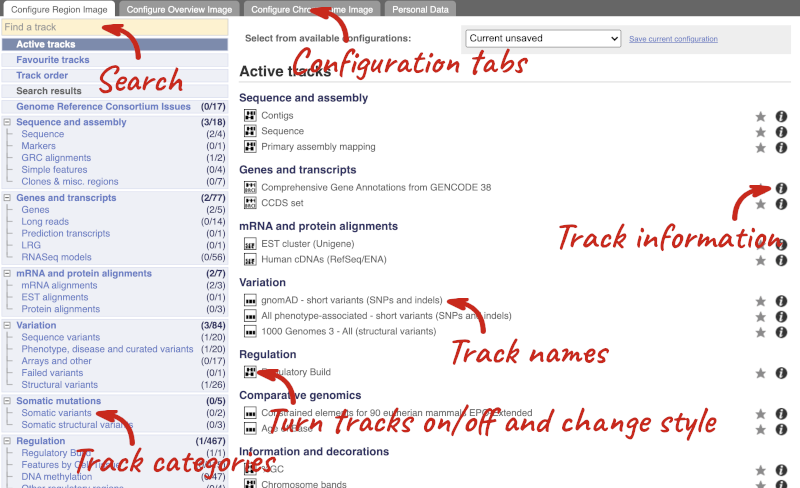
There are thousands of possible tracks that you can add. When you launch the view, you will see all the tracks that are currently turned on with their names on the left and an info icon on the right, which you can click on to expand the description of the track. Turn them on or off, or change the track style by clicking on the box next to the name. More details about the different track styles are in this FAQ: http://www.ensembl.org/Help/Faq?id=335.
You can find more tracks to add by either exploring the categories on the left, or using the Find a track option at the top left. Type in a word or phrase to find tracks with it in the track name or description.
Let’s add some tracks to this image. Add:
- Proteins (mammal) from UniProt – Labels
- 1000 Genomes - All - short variants (SNPs and indels) – Normal
Now click on the tick in the top left hand to save and close the menu. Alternatively, click anywhere outside of the menu. We can now see the tracks in the image. The proteins track is stranded, so you will see two tracks, one above and one below the contig, representing the proteins mapped to the forward and reverse strands respectively. The variants track is not stranded, so is found near the bottom of the image.
If the track is not giving you can information you need, you can easily change the way the tracks appear by hovering over the track name then the cog wheel to open a menu. To make it easier to compare information between tracks, such as spotting overlaps, you can move tracks around by clicking and dragging on the bar to the left of the track name.

Now that you’ve got the view how you want it, you might like to show something you’ve found to a colleague or collaborator. Click on the Share this page button to generate a link. Email the link to someone else, so that they can see the same view as you, including all the tracks you’ve added. These links contain the Ensembl release number, so if a new release or even assembly comes out, your link will just take you to the archive site for the release it was made on.
To return this to the default view, go to Configure this page and select Reset configuration at the bottom of the menu.
Exploring a genomic region in human
Go to Ensembl.
-
Go to the region from 32,264,000 to 32,492,000 bp on human chromosome 13. On which cytogenetic band is this region located? How many contigs make up this portion of the assembly (contigs are contiguous stretches of DNA sequence that have been assembled solely based on direct sequencing information)?
-
Zoom in on the BRCA2 gene.
-
Configure this page to turn on the LTR (repeat) track in this view. What tool was used to annotate the LTRs according to the track information? How many LTRs can you see within the BRCA2 gene? Do any overlap exons?
-
Create a Share link for this display. Email it to your neighbour. Open the link they sent you and compare. If there are differences, can you work out why?
-
Export the genomic sequence of the region you are looking at in FASTA format.
-
Turn off all tracks you added to the Region in detail page.
- Go to the Ensembl homepage, select Human from the Species drop-down list and type
13:32264000-32492000in the text box (alternatively leave the Search drop-down list as it is and type13:32264000-324920000in the text box). Click Go.This genomic region is located on cytogenetic band q13.1. It is made up of three contigs, indicated by the alternating light and dark blue coloured bars in the Contigs track.
-
Draw with your mouse a box encompassing the BRCA2 transcripts. Click on Jump to region in the pop-up menu.
- Click Configure this page in the side menu (or on the cog wheel icon in the top left hand side of the bottom image). Go into Repeats in the left-hand menu then select LTR. Click on the (i) button to find out more information.
Repeat Masker was used to annotate LTRs onto the genome.
Save and close the new configuration by clicking on ✓ (or anywhere outside the pop-up window). There are ten LTRs overlapping BRCA2, none of them overlap exons. -
Click Share this page in the side menu. Copy the URL. Get your neighbour’s email address and compose an email to them, paste the link in and send the message. When you receive the link from them, open the email and click on your link. You should be able to view the page with the new configuration and data tracks they have added to in the Location tab. You might see differences where they specified a slightly different region to you, or where they have added different tracks.
Here is the Share link from the video answer: https://may2021.archive.ensembl.org/Homo_sapiens/Share/71a173bba78f0dbe03e48d3240424943?redirect=no;mobileredirect=no
-
Click Export data in the side menu. Leave the default parameters as they are (FASTA sequence should already be selected). Click Next>. Click on Text. Note that the sequence has a header that provides information about the genome assembly (GRCh38), the chromosome, the start and end coordinates and the strand. For example:
>13_dna:chromosome_chromosome:GRCh38:13:32311910:32405865:1 - Click Configure this page in the side menu. Click Reset configuration. Click ✓.
Exploring a genomic region in mouse
Go to the Ensembl homepage.
-
Go to the region from 150,320,000 to 150,540,000 bp on mouse chromosome 5. How many contigs make up this portion of the assembly (contigs are contiguous stretches of DNA sequence that have been assembled solely based on direct sequencing information)?
-
Zoom in on the Brca2 gene.
-
Configure this page to turn on the LTR (repeat) track in this view. What tool was used to annotate the LTRs according to the track information? How many LTRs can you see within the Brca2 gene? Do any overlap exons?
-
Create a Share link for this display. Email it to your neighbour. Open the link they sent you and compare. If there are differences, can you work out why?
-
Export the genomic sequence of the region you are looking at in FASTA format.
-
Turn off all tracks you added to the Region in detail page.
- Select Mouse from the Species search list and type
5:150320000-150540000in the text box (or alternatively leave the Search drop-down list like it is and typemouse 5:150320000-150540000in the text box). Click Go.It is made up of five contigs, indicated by the alternating light and dark blue coloured bars in the Contigs track. Note the tiny contig, AEKQ02165236.1, which splits AC084217.7 in two.
-
Draw with your mouse a box encompassing the Brca2 transcripts. Click on Jump to region in the pop-up menu.
- Click Configure this page in the side menu (or on the cog wheel icon in the top left hand side of the bottom image). Go to Repeats in the left-hand menu then select LTRs (Repeats (Mouse)). Click on the (i) button to find out more information.
Repeat Masker was used to annotate LTRs onto the genome.
Save and close the new configuration by clicking on ✓ (or anywhere outside the pop-up window).
There are seven LTRs overlapping Brca2, none of them overlap exons.
-
Click Share this page in the side menu. Select the link and copy. Get your neighbour’s email address and compose an email to them, paste the link in and send the message. When you receive the link from them, open the email and click on your link. You should be able to view the page with the new configuration and data tracks they have added to in the Location tab. You might see differences where they specified a slightly different region to you, or where they have added different tracks.
-
Click Export data in the side menu. Leave the default parameters as they are. Click Next>. Click on Text.
- Click Configure this page in the side menu. Click Reset configuration. Click ✓.
Exploring a genomic region in Oryza sativa Japonica (rice)
Go to the Ensembl Plants homepage and do the following:
-
Go to the region between 405000 and 453000 on chromosome 1 in Oryza sativa Japonica.
-
Turn on the AGILENT:G2519F-015241 microarray track. Are there any oligo probes that map to this region?
-
Highlight the region around any reverse strand probes you can see. Do they map to any Ensembl transcripts?
-
Go to the Ensembl Plants homepage. Select Oryza sativa Japonica from the Species drop-down list and type
1:405000-453000. Click Go. - Click on Configure this page to open the menu. You can find the AGILENT:G2519F-015241 track under Oligo probes in the left-hand menu, or by using the Find a track box at the top right. Turn on the track as Normal then save and close the menu. As the AGILENT:G2519F-015241 track is stranded, it appears at the top and bottom of the view.
There are 5 probes mapped to this region on the positive strand and one probe on the reverse strand.
- Drag a box around the reverse strand probe then click on Mark region to highlight.
The highlighted region maps to two transcripts: Os01t0107900-02 and Os01t0107900-01
Exploring a region in Coprinopsis cinerea okayama
Go to Ensembl Fungi. Let’s try to find some information about the region from 1,400,000 to 1,425,000 in chromosome 7 in Coprinopsis cinerea okayama:
-
How many complete genes are found in this region? How many on the forward and how many on the reverse strand?
-
Zoom in on the largest gene EFI27358. How many exons does this gene have?
-
Export the genomic sequence in FASTA format for this region.
- In the Ensembl Fungi homepage, select Coprinopsis cinerea okayama from the Species search drop-down. Enter
7:1400000-1425000in the Search bar and click Go. This will send you to the Location tab. Your region of interest is indicated by a red rectangle in the 50kb view. Look at the Genes track: each block represents a different gene. Count the number of complete genes within the rectangle.There are 7 complete genes in the region.
- Look at the Region in detail view (the most detailed view at the bottom of the page). You can zoom into a region by clicking and dragging your mouse (you can change your mouse action in the top right-hand corner of the view under **Drag/Select) and selecting Jump to region in the pop-up menu. Count the number of blocks you can see for EFI27358.
The EFI27358 gene has 23 exons.
Click on the transcript ID CZT99117 in the transcript table.
It has 4 exons.
- We want to export the genomic sequence for our original region (not just the EFI27358 gene). You can reset the view by entering
7:1400000-1425000in the Location bar above the Region in detail view or hitting the Back button on your internet browser. Click on Export data in the left-hand panel. In the pop-up menu, select FASTA from the drop-down and click Next >. You can export the sequence as is (text) or as a compressed file (.gz).If you choose to download the sequence as text, your browser might open the FASTA file in a new tab. In this case, just right-click on any white space and select Save As… from the menu.
Exploring a genomic region in Salmonella enterica
Go to Ensembl Bacteria and do the following:
-
Search for the Salmonella enterica subsp. enterica serovar Typhi str. Ty2 (GCA_000007545) (Hint: type
Tyinto the Search for a genome box). -
Go to the region Chromosome:2000605-2009742.
-
How many genes are annotated in this region? How many are on the forward strand? How many are on the reverse strand?
-
Go to the Ensembl Bacteria homepage. Type
Ty2into the Search for a genome box. Click on the auto-completed genome name to navigate to the species information page. -
Type
Chromosome:2000605-2009742into the search box. Click Go. -
There are 8 genes annotated in this region, all on the reverse strand.
Genes and transcripts
You can find out lots of information about Ensembl genes and transcripts using the browser. If you’re already looking at a region view, you can click on any transcript and a pop-up menu will appear, allowing you to jump directly to that gene or transcript.
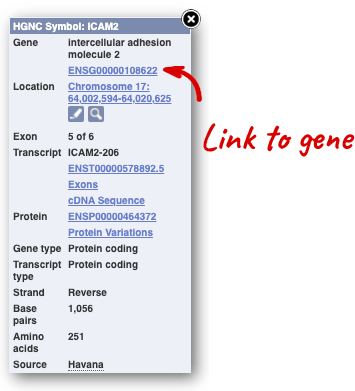
Alternatively, you can find a gene by searching for it. You can search for gene names or identifiers, and also phenotypes or functions that might be associated with the genes.
We’re going to look at the human UQCRQ gene. From ensembl.org, type UQCRQ into the search bar and click the Go button. You will get a list of hits with the human gene at the top.
Where you search for something without specifying the species, or where the ID is not restricted to a single species, the most popular species will appear first, in this case, human, mouse and zebrafish appear first. You can restrict your query to species or features of interest using the options on the left.
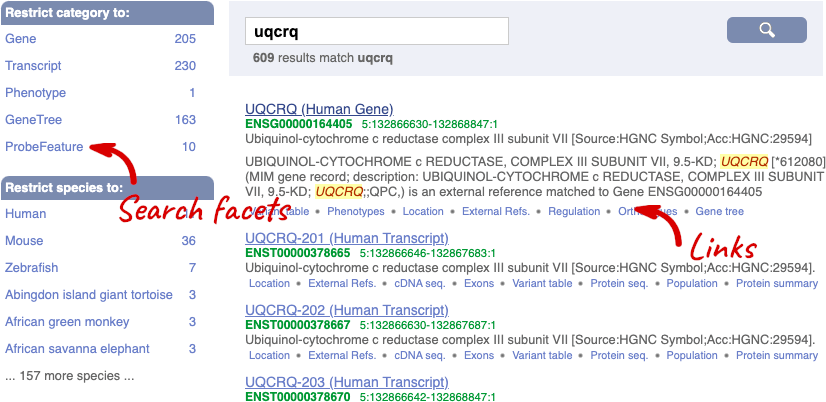
The gene tab
Click on the gene name or Ensembl ID. The Gene tab should open:
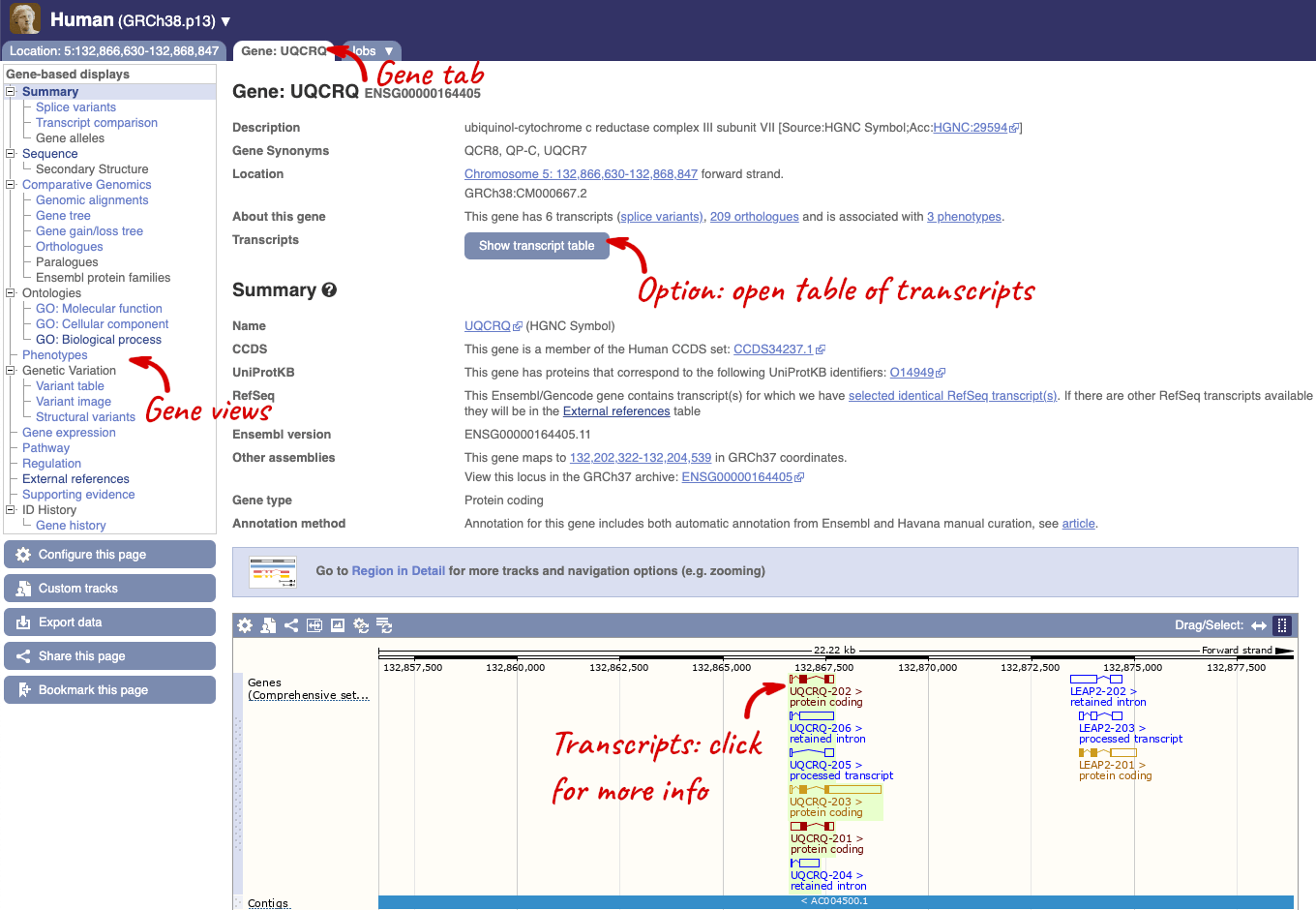
This page summarises the gene, including its location, name and equivalents in other databases. At the bottom of the page, a graphic shows a region view with the transcripts. We can see exons shown as blocks with introns as lines linking them together. Coding exons are filled, whereas non-coding exons are empty. We can also see the overlapping and neighbouring genes and other genomic features.
There are different tabs for different types of features, such as genes, transcripts or variants. These appear side-by-side across the blue bar, allowing you to jump back and forth between features of interest. Each tab has its own navigation column down the left hand side of the page, listing all the things you can see for this feature.
Let’s walk through this menu for the gene tab. How can we view the genomic sequence? Click Sequence at the left of the page.
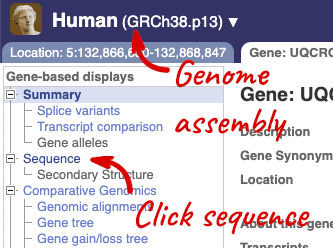
The sequence is shown in FASTA format. The FASTA header contains the genome assembly, chromosome, coordinates and strand (1 or -1) – this gene is on the positive strand.
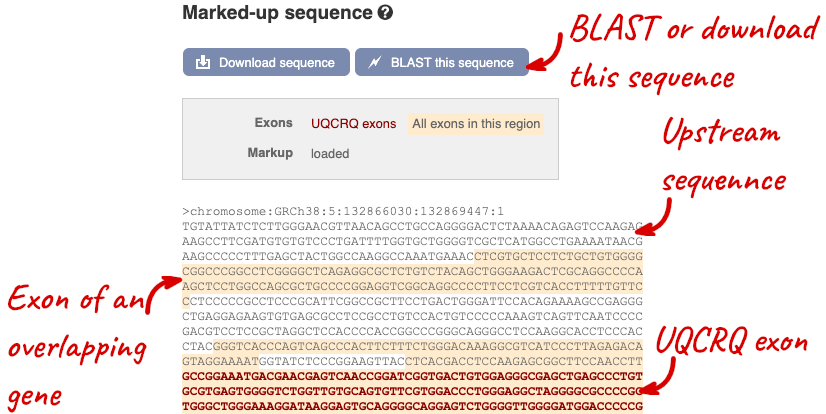
Exons are highlighted within the genomic sequence, both exons of our gene of interest and any neighbouring or overlapping gene. By default, 600 bases are shown up and downstream of the gene. We can make changes to how this sequence appears with the blue Configure this page button found at the left. This allows us to change the flanking regions, add variants, add line numbering and more. Click on it now.
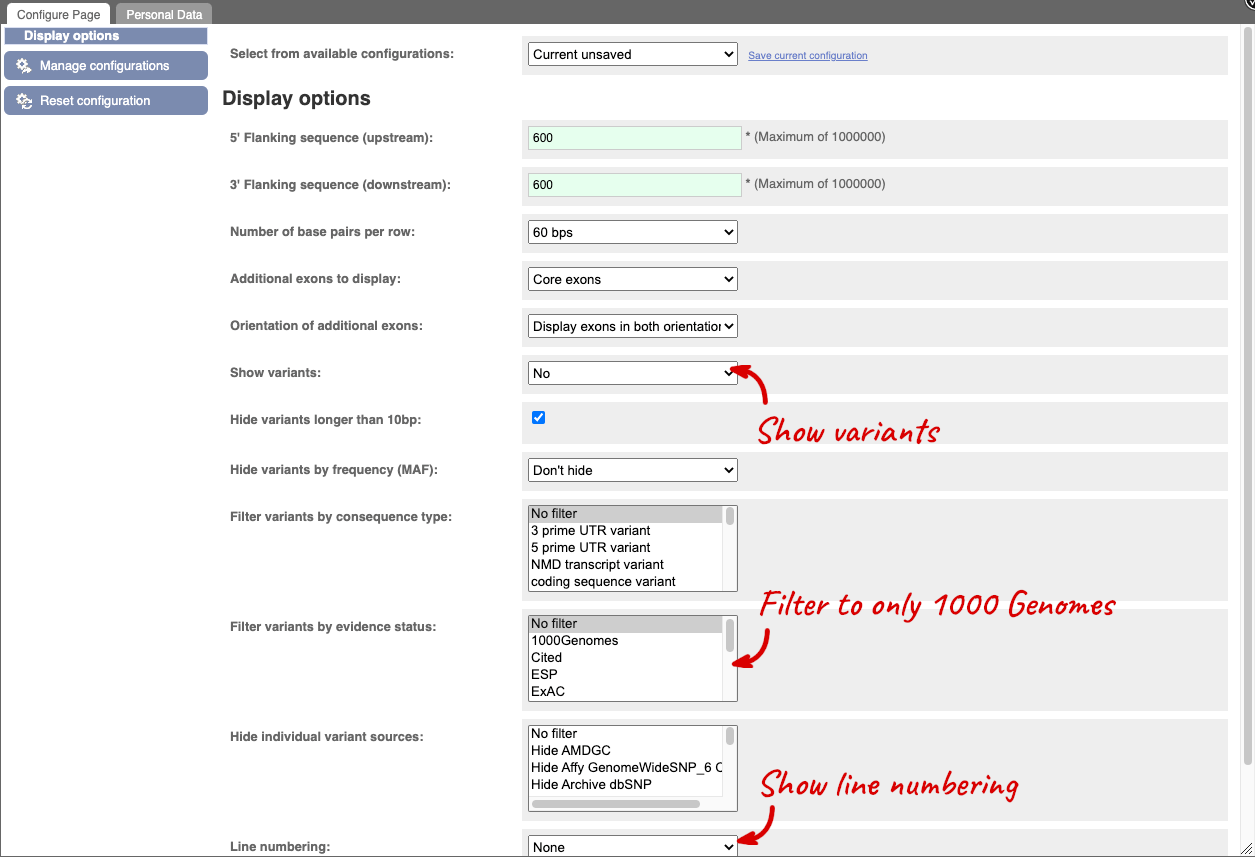
Once you have selected changes (in this example, Show variants, 1000 Genomes variants and Line numbering) click at the top right.
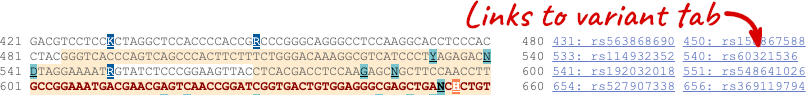
You can download this sequence by clicking in the Download sequence button above the sequence:

This will open a dialogue box that allows you to pick between plain FASTA sequence, or sequence in RTF, which includes all the coloured annotations and can be opened in a word processor. If you want run a sequence analysis tool, download as FASTA sequence, whereas if you want to analyse the sequence visually, RTF is best for this. This button is available for all sequence views.
To find out what the protein does, have a look at GO terms from the Gene Ontology consortium. There are three pages of GO terms, representing the three divisions in GO: Biological process (what the protein does), Cellular component (where the protein is) and Molecular function (how it does it). Click on GO: Biological process to see an example of the GO pages.

Here you can see the functions that have been associated with the gene. There are three-letter codes that indicate how the association was made, as well as links to the specific transcript they are linked to.
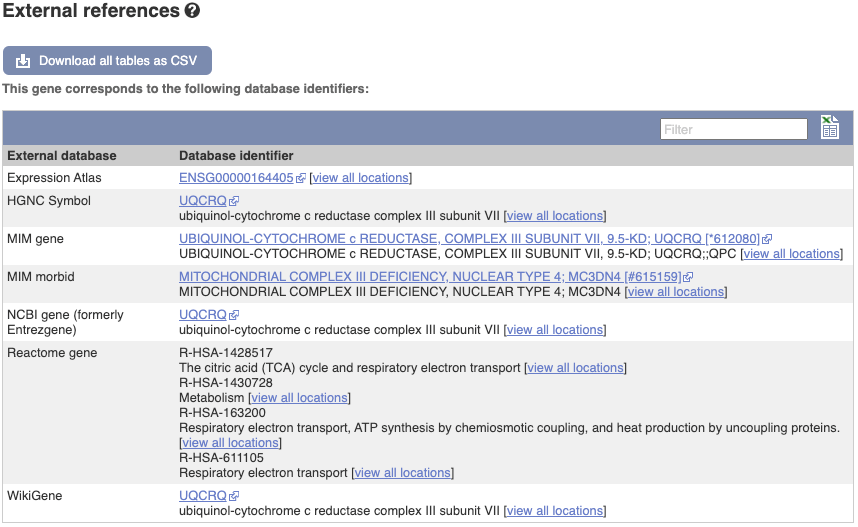
We also have links out to other databases which have information about our genes and may focus on other topics that we don’t cover, like Gene Expression Atlas or OMIM. Go up the left-hand menu to External references:
Demo: The transcript tab
We’re now going to explore the different transcripts of UQCRQ. Click on Show transcript table at the top.

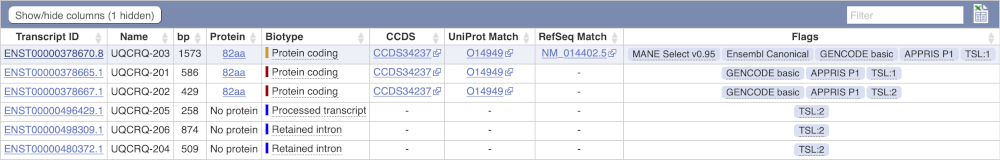
Here we can see a list of all the transcripts of UQCRQ with their identifiers, lengths, biotypes and flags to help you decide which ones to look at.
If we were to only choose one transcript to analyse, we would choose UQCRQ-203 because it is the MANE Select and Ensembl Canonical. This means it is both 100% identical to the RefSeq transcript NM_014402.5 and both Ensembl and NCBI agree that it is the most biologically important transcript.
Click on the ID, ENST00000378670.8.
You are now in the Transcript tab for UQCRQ-203. We can still see the gene tab so we can easily jump back. The left hand navigation column provides several options for the transcript UQCRQ-203 - many of these are similar to the options you see in the gene tab, but not all of them. If you can’t find the thing you’re looking for, often the solution is to switch tabs.
Click on the Exons link. This page is useful for designing RT-PCR primers because you can see the sequences of the different exons and their lengths.
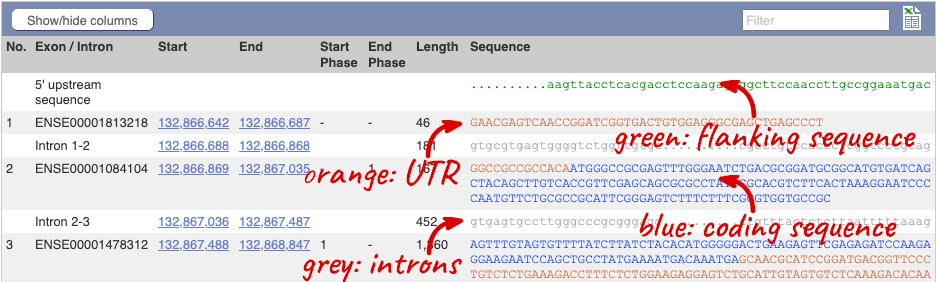
You may want to change the display (for example, to show more flanking sequence, or to show full introns). In order to do so click on Configure this page and change the display options accordingly.
Now click on the cDNA link to see the spliced transcript sequence with the amino acid sequence. This page is useful for mapping between the RNA and protein sequences, particularly genetic variants.
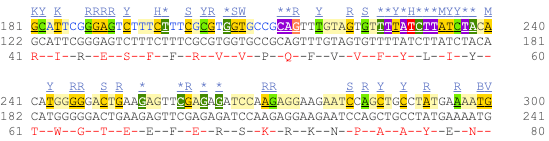
UnTranslated Regions (UTRs) are highlighted in dark yellow, codons are highlighted in light yellow, and exon sequence is shown in black or blue letters to show exon divides. Sequence variants are represented by highlighted nucleotides and clickable IUPAC codes are above the sequence.
Next, follow the General identifiers link at the left. Just like the External References page in the gene tab, this page shows links out to other databases such as RefSeq, UniProtKB, PDBe and others, this time linked to the transcript or protein product, rather than the gene.
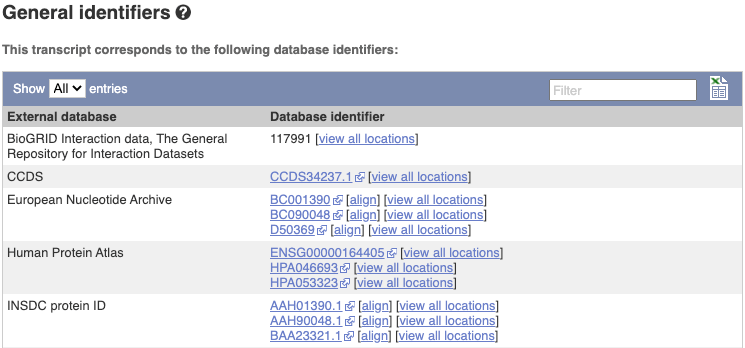
If you’re interested in protein domains, you could click on Protein summary to view domains from Pfam, PROSITE, Superfamily, InterPro, and more. These are all plotted against the transcript sequence, with the exons shown in alternating shades of purple at the top of the page. Alternatively, you can go to Domains & features to see a table of the same information.
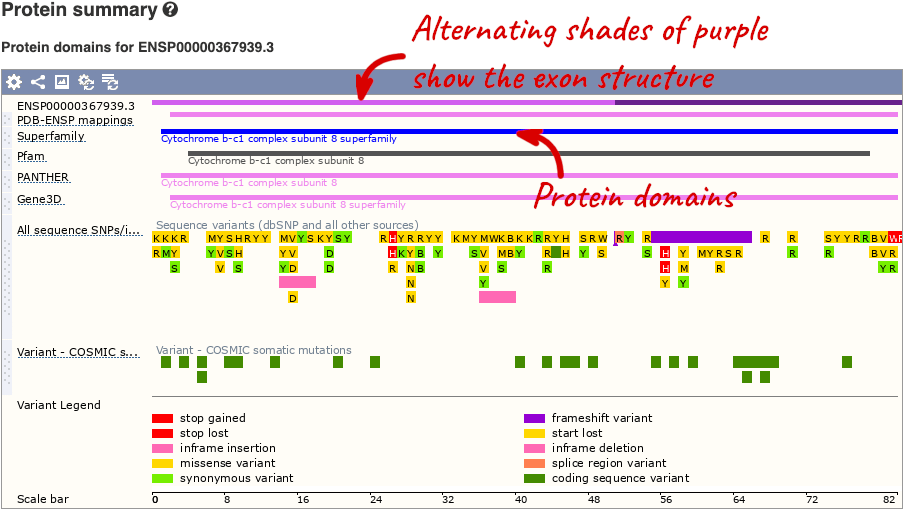

You can also see the structure of the protein from the PDB by clicking on PDB 3D Protein model.
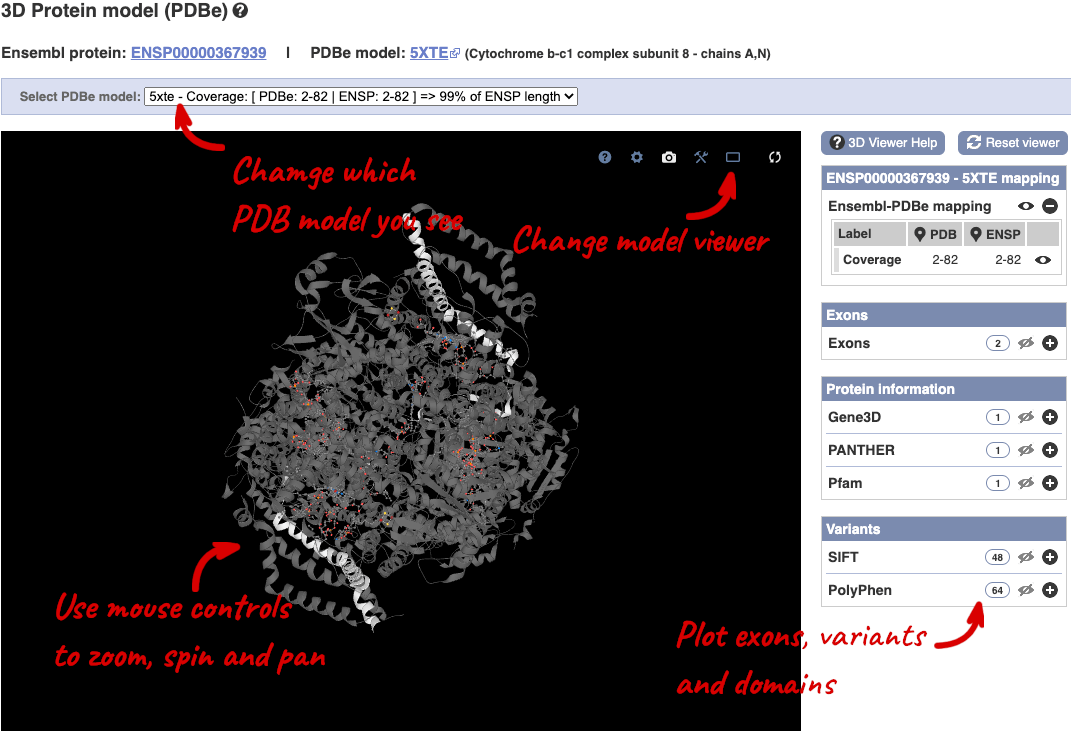
This uses LiteMol to show a 3D protein. You can use all the normal controls that you would use with LiteMol, plus plot Ensembl features like Exons and variants onto the structure using the options on the right. We allow you to see the top ten PDB models for this protein, based on coverage and quality scores, you can choose which at the top of the viewer.
Exploring the MYH9 gene in human
- In Ensembl, find the human MYH9 (myosin, heavy chain 9, non-muscle) gene and open the Gene tab.
- On which chromosome and which strand of the genome is this gene located?
- How many transcripts (splice variants) are there and how many are protein coding?
- What is the longest protein-coding transcript, and how long is the protein it encodes?
- Which transcript would you take forward for further study?
-
Click on Phenotypes at the left side of the page. Are there any diseases associated with this gene, according to Mendelian Inheritance in Man (MIM)?
-
What are some functions of MYH9 according to the Gene Ontology (GO) consortium? Have a look at the GO: Biological process pages for this gene.
- In the transcript table, click on the transcript ID for MYH9-201, and go to the Transcript tab.
- How many exons does it have?
- Are any of the exons completely or partially untranslated?
- Is there an associated sequence in UniProtKB/Swiss-Prot? Have a look at the General identifiers for this transcript.
- Are there microarray (oligo) probes that can be used to monitor ENST00000216181 expression?
- Select Human from the Species drop-down list and type
MYH9. Click Go. Click on MYH9 (Human Gene) in the search results which will send you to the Gene tab.- The gene is located on chromosome 22 on the reverse strand.
- Ensembl has 23 transcripts annotated for this gene, of which 6 are protein-coding.
- The longest protein-coding transcript is MYH9-215 and it codes for a protein that is 1,981 amino acids long.
- MYH9-201 is the transcript I would take forward for further study, as it is the MANE Select transcript (for a description, mouse-over the MANE Select flag in the transcript table).
- Click on Phenotypes in the left-hand panel to see the associated phenotypes. There is a large table of phenotypes. To see only the ones from MIM, type
MIMinto the filter box at the top right-hand corner of the table.These are some of the phenotypes associated with MYH9 according to MIM: Deafness, Autosomal dominant 17 and Macrothrombocytopenia and granulocyte inclusions with or without nephritis or sensorineural hearing loss. You can click on the records for more information.
-
The Gene Ontology project maps terms to a protein in three classes: biological process, cellular component, and molecular function. Click on GO: Biological process on the left-hand panel. Angiogenesis, cell adhesion, and protein transport are some of the roles associated with MYH9. All GO terms are associated with a single transcript: ENST00000216181.
- Click on ENST00000216181.11 in the transcript table. You should now be on the Transcript tab.
- It has 41 exons, shown in the Transcript summary.
Click on the Exons link in the left-hand panel.
- Exon 1 is completely untranslated, and exons 2 and 41 are partially untranslated (UTR sequence is shown in orange). You can also see this in the cDNA view if you click on the cDNA link in the left side menu.
Click on General identifiers in the left-hand panel.
- P35579.254 from UniProt/Swiss-Prot matches the translation of the Ensembl transcript. Click on P35579.254 to go to UniProtKB, or click align for the alignment.
- Click on Oligo probes in the left-hand panel.
Probesets from Affymetrix, Agilent, Codelink, Illumina, and Phalanx OneArray match to this transcript sequence. Expression analysis with any of these probesets would reveal information about the transcript. Hint: this information can sometimes be found in the [ArrayExpress Atlas] (https://www.ebi.ac.uk/biostudies/arrayexpress).
Finding a gene associated with a phenotype
Phenylketonuria is a genetic disorder caused by an inability to metabolise phenylalanine in any body tissue. This results in an accumulation of phenylalanine causing seizures and intellectual disability.
(a) Search for phenylketonuria from the Ensembl homepage and narrow down your search to only genes. What gene is associated with this disorder?
(b) How many protein coding transcripts does this gene have? View all of these in the transcript comparison view.
(c) What is the MIM gene identifier for this gene?
(d) Go to the MANE Select transcript and look at its 3D structure. In the model 2pah, how many protein molecules can you see?
(a) Start at the Ensembl homepage (http://www.ensembl.org).
Type phenylketonuria into the search box then click Go. Choose Gene from the left hand menu.
The gene associated with this disorder is PAH, phenylalanine hydroxylase, ENSG00000171759.
(b) If the transcript table is hidden, click on Show transcript table to see it.
There are six protein coding transcripts.
Click on Transcript comparison in the left hand menu. Click on Select transcripts. Either select all the transcripts labelled protein coding one-by-one, or click on the drop down and select Protein coding. Close the menu.
(c) Click on External references.
The MIM gene ID is 612349.
(d) Open the transcript table and click on the ID for the MANE Select: ENST00000553106.6. Go to PDB 3D protein model in the left-hand menu.
The model 2pah is shown by default. It has two protein molecules in it. You may need to rotate the model to see this clearly.
Exploring the Dpp6 gene in mouse
Genetic variation in the dipeptidylpeptidase 6 Gene (DPP6) in humans has previously been strongly associated with amyotrophic lateral sclerosis (ALS), a lethal disorder caused by progressive degeneration of motor neurons in the brain.
-
Go to the Ensembl homepage, search for the Dpp6 gene in mouse and click on the transcript ID ENSMUST00000071500 to open the transcript tab. How many exons make up this transcript?
-
Click on Exons to display the exon sequences of the transcript. Which exon contains the translation start? What is the exon ID of the largest exon? What is the start and end phase of exon 2?
-
Go to the Protein summary. How many protein domains or features fall within the second exon? What is the Pfam protein domain at the C-terminus of the protein and how many exons does it fall into? Which amino acid positions does the domain above cover?
-
Go to Domains and features. Which domains are associated with Pfam? How many genes in the mouse genome have the IPR002469 domain? What chromosomes are these genes found on?
- Select Mouse from the Species search drop-down and type
Dpp6and click Go. Click on Dpp6-201 (Mouse Transcript, Strain: reference (CL57BL6)) in the results.ENSMUST00000071500.13 consists of 26 exons.
- Click on Exons in the left-hand panel. The translation start is found in the first exon (ENSMUSE00000725552), shown in dark blue text.
The largest exon is the final exon (856 bp), which has the exon ID ENSMUSE00000773588. Exon 2 has a start and end phase of 0 and 1 respectively, which means that the codon at the start of the exon starts at the first nucleotide and the codon at the end of the exon ends at nucleotide 2. Notice that the end phase of each exon is the same as the start phase of the next exon.
- Click on Protein summary in the menu on the left hand side of the page. Alternating exons are shown on the protein as different shades of purple.
There are two predicted protein domains that fall within the second exon: a transmembrane helix and low complexity peptide sequence (Seg). You can click on the track names to find a description.
Click on a domain or feature to view further information.
The C-terminal Pfam domain is Peptidase_S9 (PF00326), which spans or partially spans seven exons, covering amino acid positions 582-787.
- Click on Domains & features.
Looking at the domains table you should notice that there are two domains associated with Pfam: PF00326 and PF00930.
Click on Display all genes with this domain next to IPR002469. This should now display the genes that have the IPR002469 domain located on the karyotype and as a table.
6 genes have this domain and they are found on chromosomes 1, 2, 5, 9 and 17.
Exploring the CCD7 gene in Arabidopsis thaliana
-
Find the Arabidopsis thaliana CCD7 gene on Ensembl Plants. On which chromosome and which strand of the genome is this gene located?
-
Where in the cell is the CCD7 protein located?
-
What is the source of the assigned gene name?
-
How many transcripts does it have? How long is its longest transcript (in bp)? How long is the protein it encodes? How many exons does it have? Are any of the exons completely or partially untranslated?
- Go to the Ensembl Plants homepage (http://plants.ensembl.org/). Select A. thaliana from the species list and type
CCD7in the search box. Click Go and click on the gene ID AT2G44990. You can find the strand orientation and the location under Summary in the Gene tab.The A. thaliana CCD7 gene is located on chromosome 2 on the forward strand.
- Click on GO: Cellular component in the left-hand panel.
The protein is located in the chloroplast and plastid.
- Click on Summary in the side menu.
The gene name is assigned and imported from NCBI gene (formerly Entrezgene).
- Click on Show transcript table.
There are 3 transcripts. The longest one is 2005 bp and the length of the encoded protein is 622 amino acids.
Click on the transcript ID AT2G44990.3 in the transcript table. You can find the number of exons in under in the summary information at the top of the page.
It has 6 exons.
Click on Sequence: Exons in the left-hand panel.
The first and last exons are partially untranslated (sequence shown in orange). This can also been seen from the fact that in the transcript diagrams on the Gene Summary and Transcript Summary pages the boxes representing the first and last exon are partially unfilled.
Exploring a bacterial gene in Clostridium sporogenes
Start in Ensembl Bacteria and select the Clostridium sporogenes (GCA_001444695) genome.
-
What is the gene name for the Glutamine synthetase gene?
-
Go to the transcript tab. How long is the transcript? How long is the protein?
-
What domains can be found in the protein product of this transcript? How many different domain prediction methods agree with each of these domains?
- From the Ensembl Bacteria homepage, select Clostridium sporogenes by beginning to write the species name and selecting the species from the auto-complete list. Type
Glutamine synthetaseand click on the gene ID ENSB:yZtlLO8Ti90y75J which will open the Summary display on the Gene tab..The gene name is
glnA. - Switch to the Transcript tab and go to the Summary display. You can find the length under Statistics underneath the transcript image.
The
glnAtranscript is 1,899 bp and the protein is 632 aa in length. - Click on either Protein Summary or Domains & features in the left hand menu to see graphically or as a table respectively.
6 protein domains were found. All of them predict a glutamine synthetase domain.
Exploring a gene in Escherichia coli
Start in Ensembl Bacteria and search for the Escherichia coli str. K-12 substr. MG1655 (GCA_000005845) genome.
-
What GO: biological process terms are associated with the Era gene?
-
How many different InterPro domains are found in the protein product of this gene?
-
What is the associated UniProt ID of the transcript?
Enter part of the name into the genome search box (e.g. MG1655) and then select the correct genome to go to the species information page.
- Enter
Erainto the search box and hit Go. Click the link in the first hit to go to the era gene page. From here, click GO: Biological process in the left-hand menu.There are three GO IDs: GO:0000028, GO:0006468, GO:0042274 and GO:0046777.
- Switch to the Transcript tab and go to Domains & features in the left-hand panel. Count the number of unique InterPro IDs in the table.
8 different InterPro domains are found in the protein product of Era.
- You can find the UniProt ID in the transcript table or under General identifiers in the left-hand panel.
The UniProtKB/Swiss-Prot ID is P06616.
BioMart
Follow these instructions to guide you through BioMart to answer the following query:
You have three questions about a set of human genes:
ESPN, MYH9, USH1C, CISD2, THRB, WHRN
(these are HGNC gene symbols. More details on the HUGO Gene Nomenclature Committee can be found on http://www.genenames.org)
- What are the NCBI Gene IDs for these genes?
- Are there associated functions from the GO (gene ontology) project that might help describe their function?
- What are their cDNA sequences?
Click on BioMart in the top header of a www.ensembl.org page to go to: www.ensembl.org/biomart/martview
You cannot choose any filters or attributes until you’ve chosen your dataset. Your dataset is the data type you’re working with. In this case we’re going to choose human genes, so pick Ensembl Genes then Human genes from the drop-downs.
Now that you’ve chosen your dataset, the filters and attributes will appear in the column on the left. You can pick these in any order and the options you pick will appear.
Click on Filters on the left to see the available filters appear on the main page. You’ll see that there are loads of categories of Filters to choose from. You can expand these by clicking on them. For our query, we’re going to expand GENE.
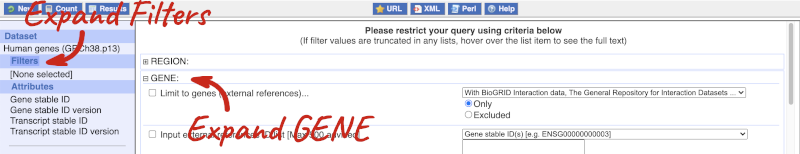
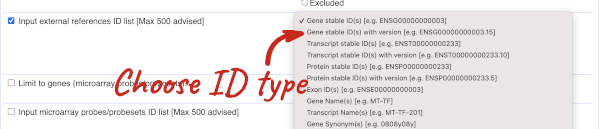
Our input data is a list of identifiers, so we’re going to use the Input external references ID list filter. This allows us to input a list of identifiers from different databases. We need to choose what kind of identifier we’re using, so that BioMart can look up the right column in a data table. You can pick these from a drop-down list, which lists the type of identifier with an example of how it looks. For our query, we have a list of gene names, so we need to pick Gene Name(s).
To check if the filters have worked, you can use the Count button at the top left, which will show you how many genes have passed the filter. If you get 0 or another number you don’t expect, this can help you to see if your query was effective.
To choose the attributes, expand this in the menu. There are six categories for human gene attributes. These categories are mutually exclusive, you cannot pick attributes from multiple categories. This means that we need to do two separate queries to get our GO terms and NCBI IDs, and to get our cDNA sequences.

The Ensembl gene and transcript IDs, with and without version numbers are selected by default. The selected attributes are also listed on the left.

We can choose the attributes we want by clicking on them. For our query, we’re going to select:
- GENE
- Gene Name
- EXTERNAL
- NCBI gene ID
- GO term accession
- GO term name
- GO term definition
We need to select the Gene Name in order to get back our original input, as this is not returned by default in BioMart. The order that you select the attributes in will define the order that the columns appear in in your output table.
You can get your results by clicking on Results at the top left.

The results table just gives you a preview of the first ten lines of your query. This allows the results to load quickly, so that if you need to make any changes to your query, you don’t waste any time. To see the full table you can click on View ## rows. You can also export the data to an xls, tsv, csv or html file. For large queries, it is recommended that you export your data as Compressed web file (notify by email), to ensure your download is not disrupted by connection issues.
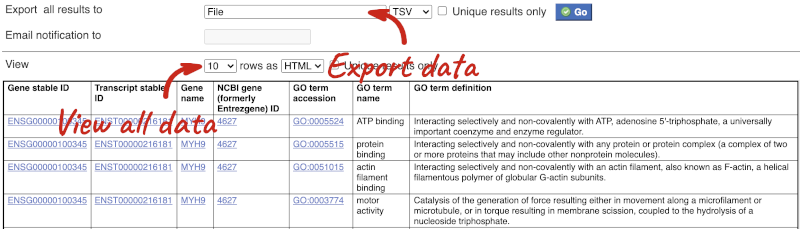
You can see multiple rows per gene in your input list, because there are multiple transcripts per gene and multiple GO terms per transcript.
To get the cDNA sequences, go back to the Attributes then select the category Sequences and expand SEQUENCES.
When you select the sequence type, the part of the transcript model you’ve chosen will be highlighted in the grpahic.
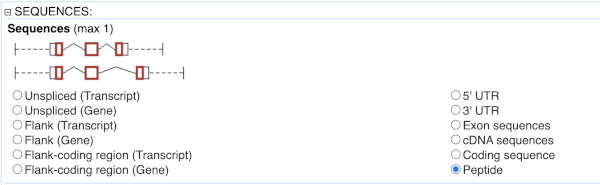
Choose cDNA sequences, then expand HEADER INFORMATION to add Gene Name to the header. Then hit Results again.

For more details on BioMart, have a look at this publication:
Kinsella, R.J. et al
Ensembl BioMarts: a hub for data retrieval across taxonomic space.
http://europepmc.org/articles/PMC3170168
Finding genes by protein domain
Find mouse proteins with Signalp cleavage sites located on chromosome 9.
As with all BioMart queries you must select the dataset, set your filters (input) and define your attributes (desired output). For this exercise: Dataset: Ensembl genes in mouse Filters: Signalp cleavage sites on chromosome 9 Attributes: Ensembl gene and transcript IDs and gene names
Go to the Ensembl homepage (http://www.ensembl.org) and click on BioMart at the top of the page. Select Ensembl genes as your database and Mouse genes as the dataset. Click on Filters on the left of the screen and expand REGION. Change the chromosome to 9. Now expand PROTEIN DOMAINS, also under filters, and select Limit to genes, choosing with With Cleavage site (Signalp) from the drop-down and then Only. Clicking on Count should reveal that you have filtered the dataset down to 217 genes.
Click on Attributes and expand GENE. Select Gene name. Now click on Results. The first 10 results are displayed by default; Display all results by selecting ALL from the drop down menu.
The output will display the Ensembl gene ID, Ensembl Transcript ID and gene names of all proteins with a Signalp cleavage site on mouse chromosome 9. If you prefer, you can also export as an Excel sheet by using the Export all results to XLS option.
Exporting homologues with BioMart
Go to Ensembl’s BioMart. For a list of Ciona savignyi Ensembl genes, export the human orthologues:
ENSCSAVG00000000002, ENSCSAVG00000000003, ENSCSAVG00000000006, ENSCSAVG00000000007, ENSCSAVG00000000009, ENSCSAVG00000000011
Do all of these genes have a homologue in human?
-
Go to BioMart (you can find a shortcut in the navigation bar at the top of any Ensemblpage) and click New. Choose the Ensembl Genes database. Choose the Ciona savignyi genes (CSAV 2.0) dataset.
-
Click on Filters in the left panel. Expand the GENE. Enter the gene list in the Input external references ID list box. Gene stable ID(s) should be preselected.
-
Click on Attributes in the left panel. Select the Homologues attributes at the top of the page. Expand the GENE section. Deselect Gene stable ID version, Transcript stable ID and Transcript stable ID version. Expand the ORTHOLOGUES [F-J] section. Select Human gene stable ID.
-
Click Results. Select View: All rows as HTML.
All but ENSCSAVG00000000006 have a homologue in human.
Convert IDs using BioMart
BioMart is a very handy tool when you want to convert IDs from different databases. The following is a list of 29 IDs of human proteins from the NCBI RefSeq database:
NP_001218, NP_203125, NP_203124, NP_203126, NP_001007233, NP_150636, NP_150635, NP_001214, NP_150637, NP_150634, NP_150649, NP_001216, NP_116787, NP_001217, NP_127463, NP_001220, NP_004338, NP_004337, NP_116786, NP_036246, NP_116756, NP_116759, NP_001221, NP_203519, NP_001073594, NP_001219, NP_001073593, NP_203520, NP_203522
Use BioMart in Ensembl to generate a list that shows to which Ensembl gene IDs and to which gene names these RefSeq IDs correspond. Do these 29 transcripts correspond to 29 genes?
-
Go to BioMart. You can find a shortcut to the tool on any Ensembl page in the navigation bar at the top of the page. Click New in the top left-hand menu if you need to start a new query. Choose the Ensembl Genes database. Choose the Human genes dataset.
-
Click on Filters in the left panel. Expand the GENE section. Select Input external references ID list - RefSeq peptide ID(s) and enter the list of IDs in the text box (either comma separated or as a list).
HINT: You may have to scroll down the menu to see these.
Count shows 10 genes (remember one gene may have multiple splice variants coding for different proteins, that is the reason why these 29 proteins do not correspond to 29 genes).
-
Click on Attributes in the left panel. Select the Features attributes page. Expand the External section. Select HGNC symbol and RefSeq Peptide ID from the External References section.
-
Click the Results button on the toolbar. Select View: All rows as HTML or export all results to a file.
Export structural variants
You can use BioMart to query variants, not just genes. (Make sure you use the right Datasets.)
(a) Export the study accession, source name, chromosome, sequence region start and end (in bp) of human structural variations (SV) on chromosome 1, starting at 130,408 and ending at 210,597.
(b) In a new BioMart query, find the alleles, phenotype descriptions, and associated genes for the human SNPs rs566014072 and rs754099015. Can you view this same information in the Ensembl browser?
(a) Choose Ensembl Variation and Human Structural Variants (GRCh38).
Filters: Region: Chromosome 1, Base pair start: 130408, Base pair end: 210597
Count shows 87 structural variants.
Attributes: Structural Variation (SV) Information: DGVa Study Accession and Source Name, Structural Variation (SV) Location: Chromosome/scaffold name, Chromosome/scaffold position start (bp) and Chromosome/scaffold position end (bp).
(b) Choose Ensembl Variation and Human Short Variation (SNPs and indels) (GRCh38).
Filters: Filter by Variation name enter: rs566014072, rs754099015
Attributes: Variant Name, Variant Alleles, Phenotype description and Associated gene.
You can view this same information in the Ensembl browser. Click on one of the variation IDs (names) in the result table. The variation tab should open in the Ensembl browser. Click Phenotype Data.
Find genes associated with array probes
Forrest et al performed a microarray analysis of peripheral blood mononuclear cell gene expression in benzene-exposed workers (Environ Health Perspect. 2005 June; 113(6): 801–807). The microarray used was the human Affymetrix U133A/B (also called U133 plus 2) GeneChip. The top 25 up-regulated probe-sets were:
207630_s_at 221840_at 219228_at 204924_at 227613_at 223454_at 228962_at 214696_at 210732_s_at 212370_at 225390_s_at 227645_at 226652_at 221641_s_at 202055_at 226743_at 228393_s_at 225120_at 218515_at 202224_at 200614_at 212014_x_at 223461_at 209835_x_at 213315_x_at
(a) Retrieve for the genes corresponding to these probe-sets the Ensembl Gene and Transcript IDs as well as their HGNC symbols and descriptions.
(b) In order to analyse these genes for possible promoter/enhancer elements, retrieve the 2000 bp upstream of the transcripts of these genes.
(c) In order to be able to study these human genes in mouse, identify their mouse orthologues. Also retrieve the genomic coordinates of these orthologues.
(a) Click New. Choose the ENSEMBL Genes database. Choose the Human genes (GRCh38) dataset.
Click on Filters in the left panel. Expand the GENE section by clicking on the + box. Select Input microarray probes/probesets ID list - AFFY HG U133 Plus 2 probe ID(s) and enter the list of probeset IDs in the text box (either comma separated or as a list).
Count shows 26 genes match this list of probes.
Click on Attributes in the left panel. Select the Features attributes page. Expand the GENE section by clicking on the + box. In addition to the default selected attributes, select Description. Expand the External section by clicking on the + box. Select HGNC symbol from the External References section and AFFY HG U133 Plus 2 probe from the Microarray probes/probesets attributes section.
Click the Results button on the toolbar. Select View All rows as HTML or export all results to a file. Tick the box Unique results only.
Your results should show that the 25 probes map to 26 Ensembl genes.
(b) Don’t change Dataset and Filters – simply click on Attributes.
Select the Sequences attributes page. Expand the SEQUENCES section by clicking on the + box. Select Flank (Transcript) and enter 2000 in the Upstream flank text box. Expand the Header information section by clicking on the + box. Select, in addition to the default selected attributes, Gene description and Gene name.
Note: Flank (Transcript) will give the flanks for all transcripts of a gene with multiple transcripts. Flank (Gene) will give the flanks for one possible transcript in a gene (the most 5’ coordinates for upstream flanking).
Click the Results button on the toolbar.
(c) You can leave the Dataset and Filters the same, and go directly to the Attributes section:
Click on Attributes in the left panel. Select the Homologues attributes page. Expand the GENE section by clicking on the + box. Select Gene name. Deselect Ensembl Transcript ID. Expand the ORTHOLOGUES [K-O] section by clicking on the + box. Select Mouse gene stable ID, Mouse chromosome/scaffold name, Mouse chromosome/scaffold start (bp) and Mouse chromosome/scaffold end (bp).
Click the Results button on the toolbar. Select View All rows as HTML or export all results to a file.
Your results should show that for most of the human genes at least one mouse orthologue has been identified.
Exporting paralogues with BioMart
Export a list of all human genes on chromosome 14 which have a paralogue, including the gene names, the last common ancestor and the identity between the genes. How many genes on chromosome 14 have a paralogue?
Go to BioMart and click New. Choose the Ensembl Genes database. Choose the Human genes dataset.
Click on Filters in the left panel. Expand the REGION section by clicking on the + box and select Chromosome/scaffold – 14. Under MULTI SPECIES COMPARISONS select Homologue filters – Paralogous Human Genes: Only. Click the Count button in the side menu.
There are 806 genes on chromosome 14 which have a paralogue.
Click on Attributes in the left panel. Select Homologues from the six options at the top. Expand the GENE section by clicking on the + box. Deselect Transcript stable ID and Transcript stable ID version and select Gene name. Under PARALOGUES select Human paralogue gene stable ID, Human paralogue associated gene name, Paralogue last common ancestor with Human, Paralogue %id. target Human gene identical to query gene and Paralogue %id. query gene identical to target Human gene. Click the Results button on the toolbar. Select View: All rows as HTML or Export all results to a File.
Exporting regulatory features with BioMart
Using the Human Regulatory Features dataset, export a list of all enhancers falling in cytogenetic band q13.2 on chromosome 22 and their activity in Aorta. How many of them are active?
Go to BioMart and click New. Choose the Ensembl Regulation database. Choose the Human Regulatory Features dataset.
Click on Filters in the left panel. Expand the REGULATORY FEATURES section by clicking on the + box and select the following:
- Chromosome - 22
- Karyotype band: Band start – q13.2, Band end - q13.2
- Feature Type – Enhancer
- Epigenome name – aorta
Click on Attributes in the left panel. Select Chromosome/scaffold name, Start (bp), End (bp), Feature type, Regulatory stable ID, Activity and Epigenome name. Click the Results button to see the results table. Select View: All and choose to see Unique results only.
There is only one enhancer active in aorta in this cytogenetic band: ENSR00001239875.
Exporting histone modification sites with BioMart
Using the Human Regulatory Evidence dataset, export a list of all H3K9me3 modified loci on chromosome Y in Aorta. What is the source of this evidence?
Go to BioMart and click New. Choose the Ensembl Regulation database. Choose the Human Regulatory Evidence dataset.
Click on Filters in the left panel. Expand the REGULATORY EVIDENCE section by clicking on the + box and select Chromosome - Y, Feature Type – H3K9me3, and Epigenome – aorta.
Click on Attributes in the left panel. Select Chromosome/scaffold name, Start (bp), End (bp), Feature type, Epigenome name and Project name. Click the Results button to see the results table. Select View: All rows as HTML or Export all results to a File.
This data comes from the Roadmap Epigenomics.







