Filter Events by Year
ESHG: Using the Ensembl Variant Effect Predictor (VEP)
Course Details
- Lead Trainer
- Aleena Mushtaq
- Event Dates
- 2022-06-11 until 2022-06-14
- Location
- Virtual
- Description
- This workshop will guide you through the Ensembl/GENCODE and MANE annotation process, allowing you to experience the challenges involved in annotation and transcript selection. You will also learn how to use the Ensembl VEP to map genetic variants onto Ensembl/GENCODE transcripts and other Ensembl annotation including regulatory regions and regions of evolutionary conservation to determine their likely functional effects and filter to prioritise variants
Demos and exercises
Variation
In any of the sequence views shown in the Gene and Transcript tabs, you can view variants on the sequence. You can do this by clicking on Configure this page from any of these views.
Let’s take a look at the Gene sequence view for HBB in human. Search for HBB and go to the Sequence view.
If you can’t see variants marked on this view, click on Configure this page and select Show variants: Yes and show links. You may also wish to add a filter to the variants to allow them to load more quickly, we’ll add Filter variants by evidence status: 1000Genomes.
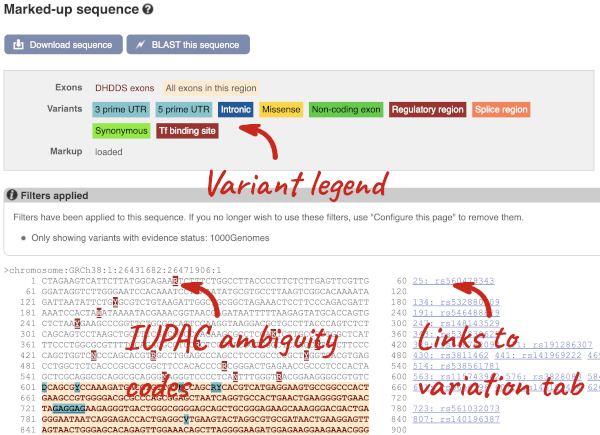
Find out more about a variant by clicking on it.
You can add variants to all other sequence views in the same way.
You can go to the Variation tab by clicking on the variant ID. For now, we’ll explore more ways of finding variants.
To view all the sequence variants in table form, click the Variant table link at the left of the gene tab.
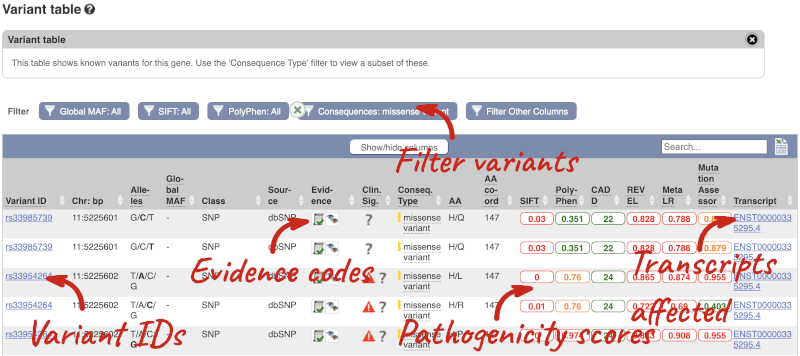
You can filter the table to only show the variants you’re interested in. For example, click on Consequences: All, then select the variant consequences you’re interested in. For display purposes, the table above has already been filtered to only show missense variants.
You can also filter by the different pathogenicity scores and MAF, or click on Filter other columns for filtering by other columns such as Evidence or Class.
The table contains lots of information about the variants. You can click on the IDs here to go to the Variation tab too.
You can also see the phenotypes associated with a gene. Click on Phenotype in the left hand menu.
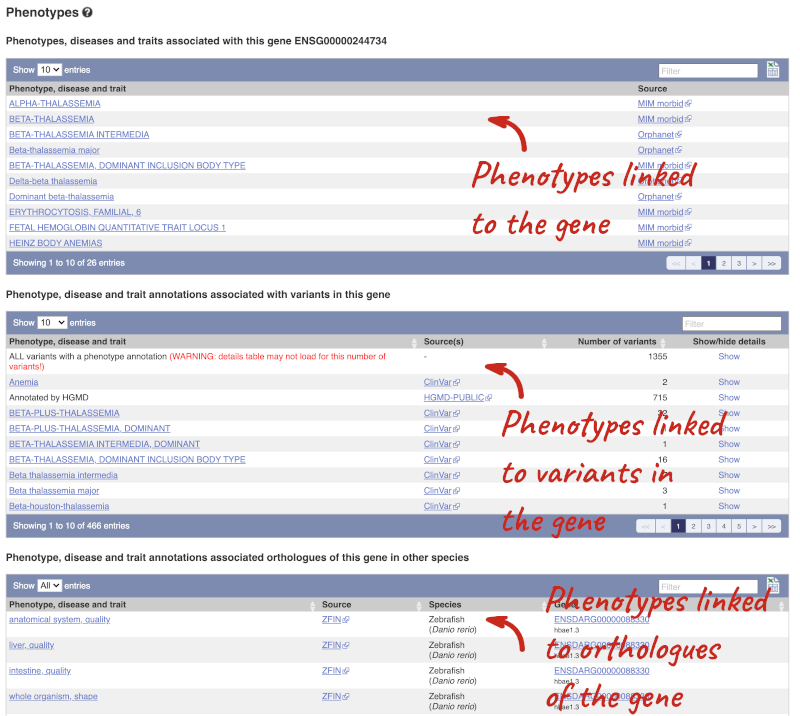
Open the transcript table and go to HBB-201 ENST00000335295, then click on Haplotypes in the left hand menu.

The Haplotypes view in the transcript tab shows you the actual protein and CDS sequences in 1000 Genomes individuals. This is possible because the 1000 Genomes study has phased genotypes, so we know which alleles occur on which of the chromosome pairs. The table lists all the versions of the protein that occur along with their frequencies, including the reference sequence and sequences with one or more alternative alleles.
Click on one of the haplotypes, we’ll go for 18K>*,19del{130}, to find out more about it. Here you will see the frequency in the 1000 Genomes subpopulations, the sequence and the 1000 Genomes individuals where this protein is found.
Let’s have a look at variants in the Location tab. Click on the Location tab in the top bar.
Configure this page and open Variation from the left-hand menu.
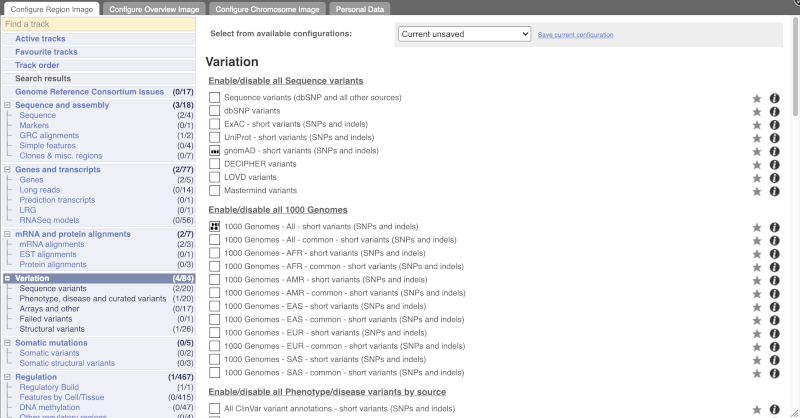
There are various options for turning on variants. You can turn on variants by source, by frequency, presence of a phenotype or by individual genome they were isolated from. You can also turn on genotyping chips.
Let’s have a look at a specific variant. If we zoomed in we could see the variant rs334 in this region, however it’s easier to find if we put rs334 into the search box. Click through to open the Variation tab.
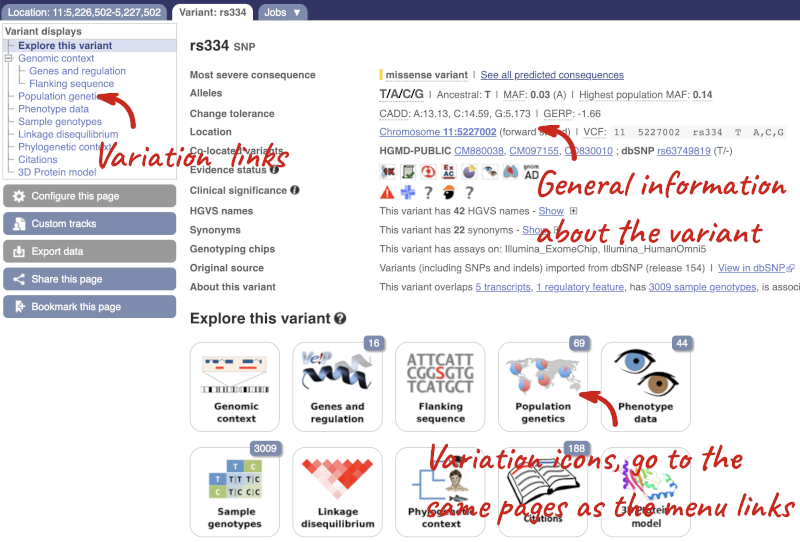
The icons show you what information is available for this variant. Click on Genes and regulation, or follow the link on the left.
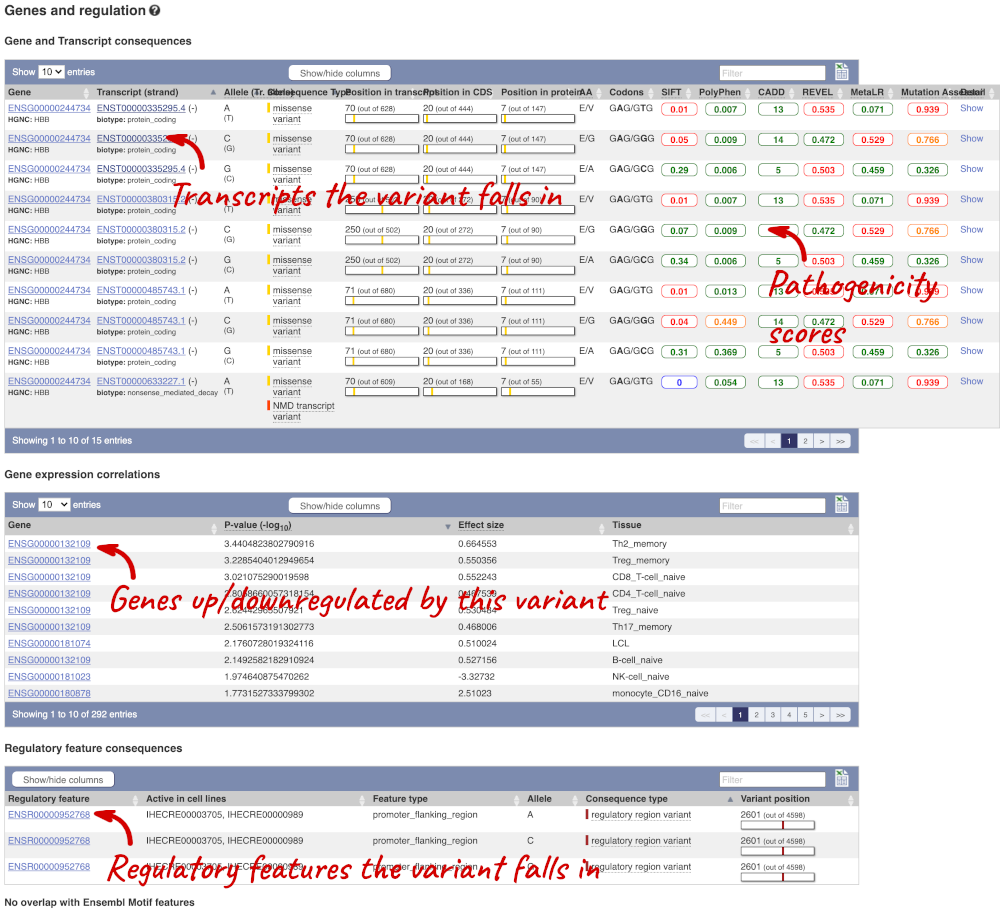
This page illustrates the genes the variant falls within and the consequences on those genes, including pathogenicity predictors. It also shows data from GTEx on genes that have increased/decreased expression in individuals with this variant, in different tissues. Finally, regulatory features and motifs that the variant falls within are shown.
We can also see the variant in the protein structure by clicking on 3D Protein model.
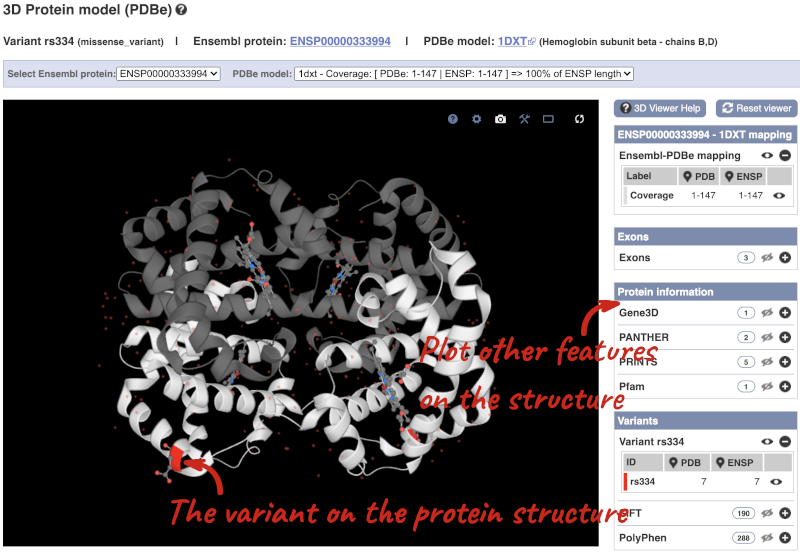
This is a LiteMol viewer, where you can rotate and zoom in on the structure. The variant location is highlighted, so you can see where it lands within the structure.
Let’s look at population genetics. Click on Population genetics in the left-hand menu.
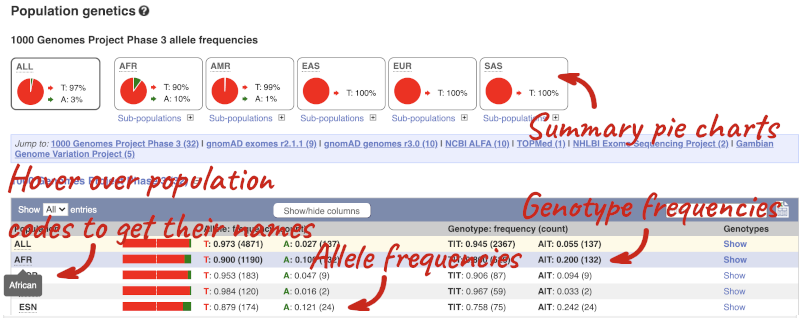
The population allele frequencies are shown by study, including 1000 Genomes and gnomAD. Where genotype frequencies are available, these are shown in the tables.
There are big differences in allele frequencies between populations. Let’s have a look at the phenotypes associated with this variant to see if they are known to be specific to certain human populations. Click on Phenotype Data in the left-hand menu.
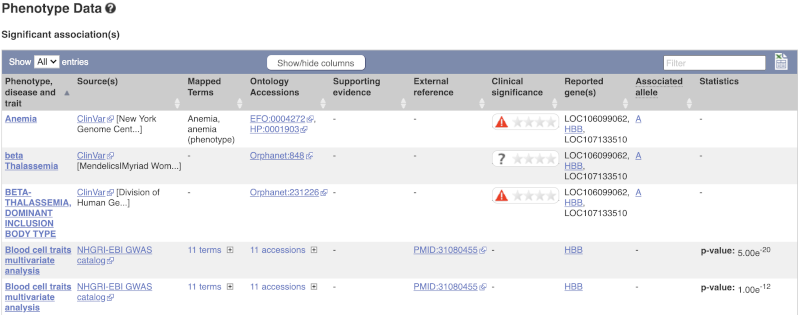
This variant is associated with various phenotypes, including sickle cell and malaria resistance. These phenotype associations come from sources including the GWAS catalog, ClinVar, Orphanet and OMIM. Where available, there are links to the original paper that made the association, the allele that is associated with the phenotype and p-values and other statistics.
Human population genetics and phenotype data
The SNP rs1738074 in the 5’ UTR of the human TAGAP gene has been identified as a genetic risk factor for a few diseases. Use Ensembl to answer the following questions:
-
In which transcripts is this SNP found?
-
What is the least frequent genotype for this SNP in the Yoruba (YRI) population from the 1000 Genomes phase 3?
-
What is the ancestral allele? Is it conserved in the 91 eutherian mammals EPO-Extended?
-
With which diseases is this SNP associated? Are there any known risk (or associated) alleles?
- Please note there is more than one way to get this answer. Either go to the Variation table of the human TAGAP gene, and use the Consequence filter to only include 5’UTR variants, or search Ensembl for
rs1738074directly. Once you’re in the Variant tab, click on Genes and regulation in the menu.This SNP is found in four transcripts of TAGAP. It is also an intron_variant to one lncRNA transcript of TAGAP-AS1.
- Click on Population genetics in the left-hand panel, or click on Explore this variant in the left-hand panel and click the Population genetics icon.
In Yoruba (YRI), the least frequent genotype is CC at the frequency of 5.6%.
- Click on Phylogenetic context in the left-hand panel.
The ancestral allele is T and it’s inferred from the alignment in primates.
Click on Select an alignment which will open a pop-up menu. Open Multiple alignments and select 91 eutherian mammals EPO-Extended. Click on Apply at the bottom of the menu to save your settings.
A region containing the SNP (highlighted in red and placed in the centre) and its flanking sequence are displayed. The T allele is conserved in all but two of the eutherian mammals displayed.
- Click Phenotype data in the left-hand panel.
This variation is associated with multiple sclerosis, celiac disease and white blood cell count. There are known risk alleles for all three diseases and the corresponding P values are provided. The allele A is associated with celiac disease. Note that the alleles reported by Ensembl are T/C. Ensembl reports alleles on the forward strand. This suggests that A was reported on the reverse strand in the original paper. Similarly, one of the alleles reported for Multiple sclerosis is G.
Exploring VNTR in human
Variable number tandem repeats (VNTRs) show high variation in the number of repeats in the population and are commonly used in forensics (DNA fingerprinting) and to study genetic diversity. (a) Go to the region from 3074666 to 3075100 bp on human chromosome 4. Which gene does it overlap? Which exon of this gene falls in this region?
(b) Configure this page to turn on Repeats (low), Simple repeats (Repeats (low)) and Tandem repeats (TRF) tracks in this view. Can you see any repeats in this exon? What tools were used to annotate the repeats according to the track information?
(c) Zoom in on the (CAG)n to see its sequence. How many CAG repeats can you see in the human reference assembly? Does this track overlap any phenotype-associated variants? What is the identifier of this variant?
(d) Go to the variant tab of the phenotype-associated variant. What is the consequence ontology of this variant? Does the reference allele match the number of repeats you have just counted? What is the shortest and longest allele?
(a) Select Search: Human and type 4:3074666-3075100 in the text box (or alternatively type human 4:3074666-3075100 in the text box). Click Go.
Click on the golden transcript falling in this region. You can see it’s exon 1 of 67 of the huntingtin gene (HTT).
(b) Click Configure this page in the side menu then select: Repeats (low), Simple repeats (Repeats (low)) and Tandem repeats (TRF).
There are three tandem repeats in this exon, and two simple repeats (low); (CAG)n and (CCG)n. Click on the track names to find more about the tools used for annotation: RepeatMasker and Tandem Repeats Finder.
(c) Draw with your mouse a box around the (CAG)n repeat. Click on Jump to region in the pop-up menu.
There are 19 CAG repeats in the human reference sequence overlapping rs71180116 indicated by a pink bar in the All phenotype-associated - short variants (SNPs and indels) track.
(d) Click on the rs71180116 ID to go to the variant tab. You can see in the summary page that this variant is classified as an inframe insertion. Either click + to show all of the alleles in the summary page or go to the Genes and regulation table. This variant has many alternative alleles which differ in the number of repeats. The first allele in the expanded Alleles section of the summary page or the first allele in the Codons column in the Genes and regulation table is the reference allele. It is composed of 19 CAG repeats just as in the Region in detail view. The shortest allele has 7 repeats, the longest has 55 repeats.
Exploring a SNP in mouse
In the paper “Altered metabolic signature in pre-diabetic NOD mice” (PloS One. 2012; 7(4): e35445), Madsen et al. have described several regulatory and coding SNPs, some of them in genes involved in ATP and adenosine metabolism, leading to potentially faulty metabolism of ATP and adenosine. The authors describe that one of the identified SNPs in the murine Entpd2 gene (rs28232063) would lead to increased amounts of available ATP, an immune activator, causing increased cell activation and possibly autoreactive T-cell activation. Use Ensembl to answer the following questions:
-
Where is the SNP located (chromosome and coordinates)?
-
What is the HGVS recommendation nomenclature for this SNP?
-
Why does Ensembl put the G allele first (G/A)?
-
Are there differences between the genotypes reported in C57BL/6NJ and NOD/ShiLtJ, according to the Mouse Genomes Project?
- From the Ensembl homepage, select Mouse from the Species search drop-down and enter
rs28232063in the search box.SNP rs28232063 is located on 2:25288362. In Ensembl, its alleles are provided relative to the forward strand.
- Click on Show under HGVS names to reveal information about HGVS nomenclature.
This SNP has got four HGVS names, one at the genomic DNA level (NC_000068.8:g.25288362G>A), two at the transcript level (ENSMUST00000148859.2:n.444-182G>A and ENSMUST00000028328.3:c.446G>A) and one at the protein level (ENSMUSP00000028328.3:p.Arg149Gln).
- In Ensembl, the allele that is present in the reference genome assembly is always put first.
G is the allele for the reference mouse genome strain C57BL/6J
- Click on Sample genotypes is the left-hand panel. The table shows genotypes reported for different mouse strains from the Mouse Genomes Project.
There are indeed differences between the genotypes reported in those two different strains. The genotype reported in C57BL/6NJ is G/G whereas in NOD/ShiLtJ the genotype is A/A.
Variation data in tomato
-
Go to Ensembl Plants and find the Solyc02g084570.3 gene in Solanum lycopersicum (tomato) and go to its Location tab. Can you see the variation track?
-
Zoom in around the last exon of this gene. What are the different types of variants seen in that region? Are any splice region variants mapped in the region? If so, what is/are the coordinate(s)?
- Select Solanum lycopersicum from the Species search drop-down menu and search for
Solyc02g084570.3. In the results page, you can click on the coordinates 2:48284598-48288482 to go straight to the Location tab. Scroll down to the Region in detail view. The variation track is shown at the bottom of the view.If you don’t see the Variation - All sources track, click Configure this page on the left-hand panel, search for the track in the pop-up menu and enable the track by clicking on the square next to the track name. Close the pop-up window and wait for the track to load.
- Zoom in around the last exon of this gene by drawing a box in the respective region (you can change your mouse action by clicking the Drag/Select icons at the top right-hand corner of the view). Note the gene is on the reverse strand (this is signified by the < sign next to the transcript name, and it is located below the Contigs track), so the last exon will be on the left hand side of that image. The variation legend is shown at the bottom of the page, telling you what the colours mean.
The types of variants seen in that region are 3’ UTR, missense, synonymous and splice region variants.
Splice region variants are shown in orange. Click on the variants to get additional information on that variant including location. You can zoom into the region if the variant block is too small to click.
The variants are found at 2:48285642 and 2:48285640-48285641. Note that the two variants overlap: one is a SNP and the other is an indel. SNPs are tagged with ambiguity codes (zoom into the region if you cannot see this). You can find a useful IUPAC ambiguity code guid on the bioinformatics.org website. Single-letter ambiguity codes are given when two or more possible nucleotides may be represented at a single base locus.
Variation data in Fusarium oxysporum
-
How many species in Ensembl Fungi have variation data?
-
Select Fusarium oxysporum (FO2) and search for the FOXG_13574T0 gene. One of its upstream variants is SNP tmp_10_6610. What are the possible alleles for this polymorphic position? Which one is on the reference genome?
-
What is the most frequent allele at this position?
-
Which samples have the genotypes C|T and T|T?
- Go to Ensembl Fungi, click on View full list of all species. You can sort the table by column. Click on the Variation database column to sort the table by species with variation data.
The table shows that we have 8 fungi species currently with variation databases.
- Click on Fusarium oxysporum in the table and on the species page search for
FOXG_13574T0. From the Gene tab, click on Variant table in the left-hand panel. You can use the filter at the top right-hand corner of the tabletmp_10_6610.The alleles are C/T, where C is the reference allele.
- Click on tmp_10_6610 in the table to open the Variant tab. Then click on Genotype frequency from the menu on the left-hand side of the page.
The most frequent allele at this position is C with a frequency of 0.850.
- Click on Sample genotypes in the menu on the left.
The table shows that sample 909454 has the C|T genotype and 909455 has the T|T genotype.
VEP
We have identified 10 variants on human chromosome eight in a patient with neurodegenerative symptoms including dystonia.
We will use the Ensembl VEP to determine:
- Have my variants already been annotated in Ensembl?
- What genes are affected by my variants?
- Do any of my variants affect gene regulation?
Go to the front page of Ensembl and click on the Variant Effect Predictor.

This page contains information about the VEP, including links to download the script version of the tool. Click on Launch VEP to open the input form:
The data is in VCF format:
chromosome coordinate id reference alternative
Put the following into the Paste data box:
8 22410000 var1 G A
8 22412087 var2 C G
8 22408305 var3 T C
8 22409953 var4 C T
8 22414115 var5 C T
8 22406904 var6 GA C
8 22366942 var7 G A
8 22408348 var8 C T
8 22404716 var9 GCTGCTGCTGCTGC GCTGCTGC
8 22404808 var10 T C
The VEP will automatically detect that the data is in VCF.
There are further options that you can choose for your output. These are categorised as Identifiers, Variants and frequency data, Additional annotations, Predictions, Filtering options and Advanced options. Let’s open all the menus and take a look.
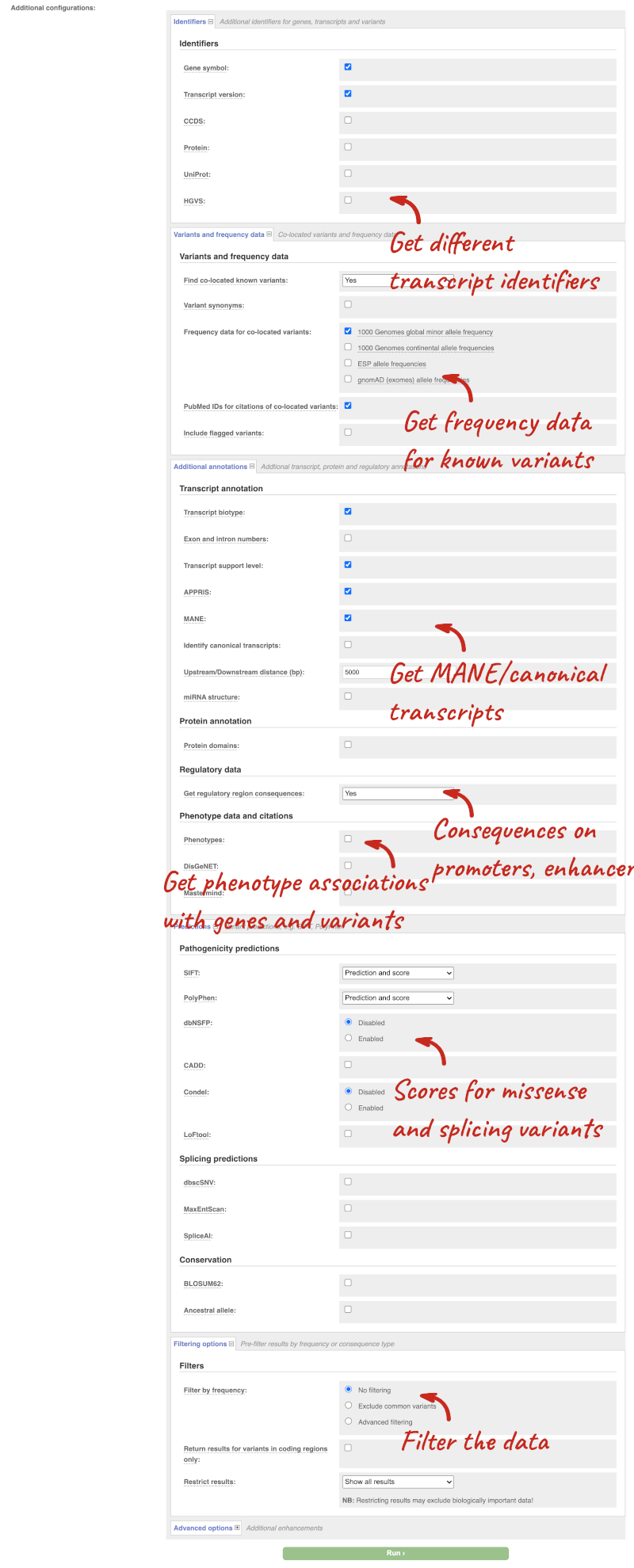
Hover over the options to see definitions.
We’re going to select some options:
- HGVS, annotation of variants in terms of the transcripts and proteins they affect, commonly-used by the clinical community
- gnomAD (exomes) allele frequencies
- Protein matches
- Phenotypes
When you have selected everything you need, scroll right to the bottom and click Run.

The display will show you the status of your job. It will say Queued, then automatically switch to Done when the job is done, you do not need to refresh the page. You can edit or discard your job at this time. If you have submitted multiple jobs, they will all appear here.
Click View results once your job is done.
In your results you will see a graphical summary of your data, as well as a table of your results.
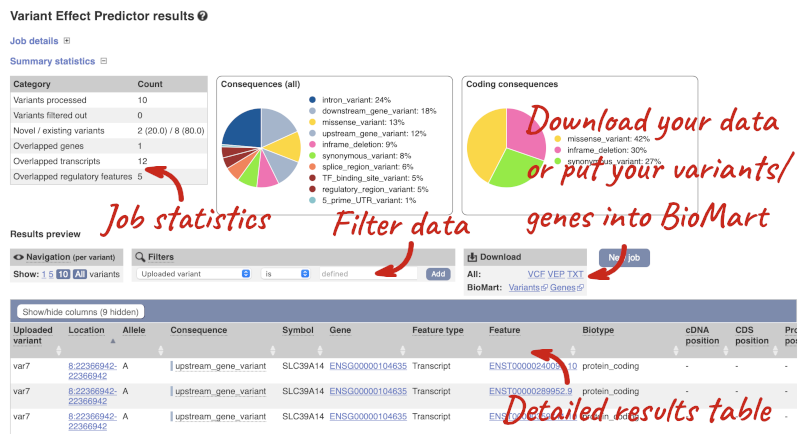
The results table is enormous and detailed, so we’re going to go through it by section. The first column is Uploaded variant. If your input data contains IDs, like ours does, the ID is listed here. If your input data is only loci, this column will contain the locus and alleles of the variant. You will notice that the variants are not neccessarily in the order they were in your input. You will also see that there are multiple lines in the table for each variant, with each line representing one transcript or other feature the variant affects.
You can mouse-over any column name to get a definition of what is shown.
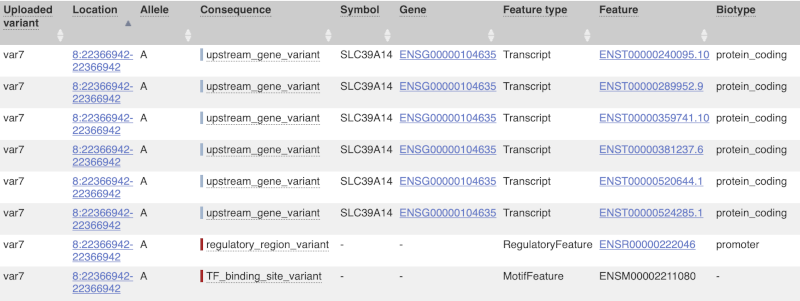
The next few columns give the information about the feature the variant affects, including the consequence. Where the feature is a transcript, you will see the gene symbol and stable ID and the transcript stable ID and biotype. Where the feature is a regulatory feature, you will get the stable ID and type. For a transcription factor binding motif (labelled as a Motif name) you will see just the ID. Most of the IDs are links to take you to the gene, transcript or regulatory feature homepage.
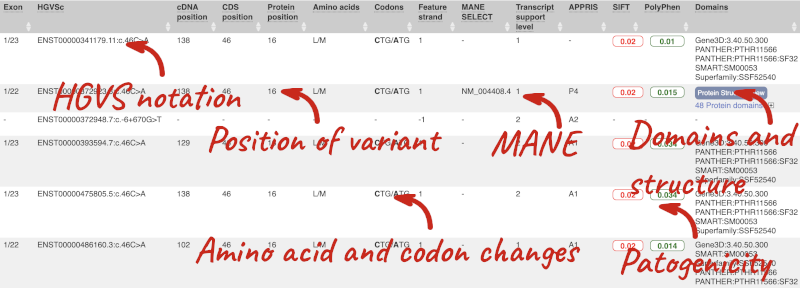
This is followed by details on the effects on transcripts, including the position of the variant in terms of the exon number, cDNA, CDS and protein, the amino acid and codon change, transcript flags, such as MANE, on the transcript, which can be used to choose a single transcript for variant reporting, and pathogenicity scores. The pathogenicity scores are shown as numbers with coloured highlights to indicate the prediction, and you can mouse-over the scores to get the prediction in words. Two options that we selected in the input form are the HGVS notation, which is shown to the left of the image below and can be used for reporting, and the Protein matches to the right. The Protein matches list the proteins domains found, and where available, provide a link to the 3D protein model which will launch a LiteMol viewer, highlighting the variant position.
Where the variant is known, the ID of the existing variant is listed, with a link out to the variant homepage. In this example, IDs from dbSNP and COSMIC are shown, but sometimes you will see IDs from other sources as well. The VEP also looks up the variant in the Ensembl database and pulls back the allele frequency (AF in the table), which will give you the 1000 Genomes Global Allele Frequency and allele frequencies from gnomAD. We can also see ClinVar clinical significance and the phenotypes associated with known variants or with the genes affected by the variants, with the variant ID listed for variant associations and the gene ID listed for gene associations, along with the source of the association.
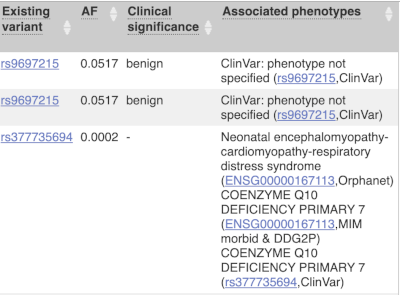
For variants that affect transcription factor binding motifs, there are columns that show the effect on motifs (you may need to click on Show/hide columns at the top left of the table to display these). Here you can see the position of the variant in the motif, if the change increases or decreases the binding affinity of the motif and the transcription factors that bind the motif.
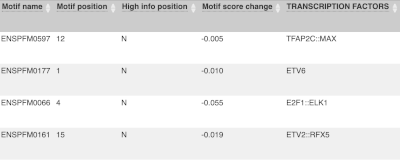
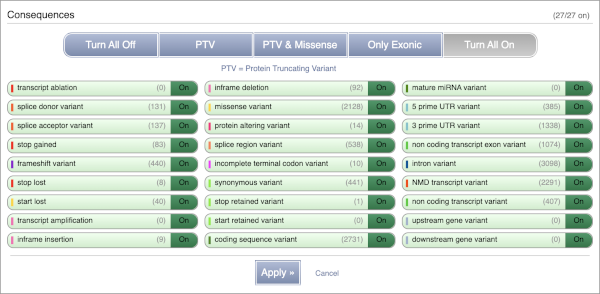
Let’s sort the table by Impact using the little upward triangle next to the column name (you may need to click on Show/hide columns to find it).
Impact is a subjective classification of the severity of the variant consequence, based on agreement with other variant annotators. It groups variant consequences into four categories: high, moderate, low and modifier. There is no high impact variant on our list, but we have some moderate variant classes such as missense or inframe deletion. Let’s filter the table to get rid of low impact variants now.
Above the table is the Filter option, which allows you to filter by any column in the table. You can select a column from the drop-down, then a logic option from the next drop-down, then type in your filter in the following box. We will try a filter of Impact, followed by is then moderate, which will give us only variant consequences, which might change protein effectiveness. You’ll notice that as you type moderate, the VEP will make suggestions for an autocomplete.
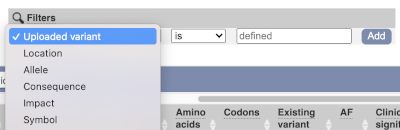
We are still left with quite a few variants. As we are dealing with a rare disorder, we can filter them by the allele frequency assuming that varaints causal of rare diseases should be very rare in the population. Choose gnomADe AF as the filter now, followed by < then 0.005, which will leave us with rare variants found below 0.5% frequency in the general population.
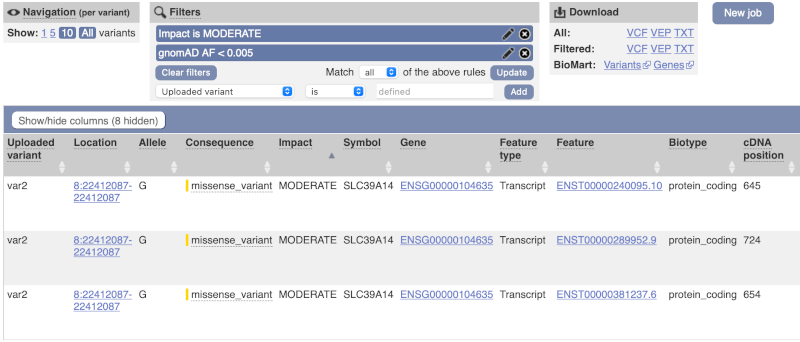
Only one variant meets our criteria. It is a benign missense variant reported on three different transcripts of the SLC39A14 gene, including the MANE Select transcript (NM_001128431.4). Note that the allele frequency for some rare or novel variants might be unknown. Let’s take that into account by editing our allele frequency filter. Change the gnomADe AF filter to is not then defined (leave the text box empty to set it to default defined).
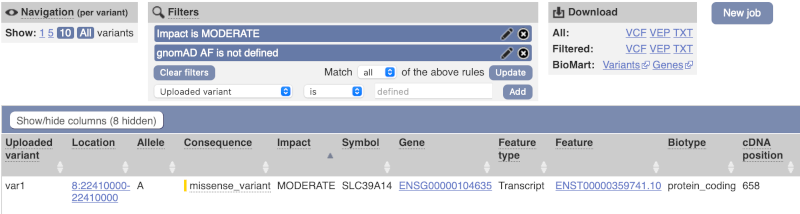
We are left with one pathogenic missense variant of unknown frequency reported on the MANE Plus clinical transcript (NM_015359.6) and associated with hypermagnesemia with dystonia, which can potentially explain our clinical case.
You can export your VEP results in various formats, including VCF. When you export as VCF, you will get all the VEP annotation listed under CSQ in the INFO column. After filtering your data, you will see that you have the option to export only the filtered data. You can also drop all the genes you have found into the Gene BioMart, or all the known variants into the Variation BioMart to export further information about them.
Running CFTR variants through VEP
Resequencing of the genomic region of the human CFTR (cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) gene (ENSG00000001626) has revealed the following variants. The alleles defined in the forward strand:
- G/A at 7: 117,530,985
- T/C at 7: 117,531,038
- T/C at 7: 117,531,068
Use the VEP tool in Ensembl and choose the options to see SIFT and PolyPhen predictions. Do these variants result in a change in the proteins encoded by any of the Ensembl genes? Which gene? Have the variants already been found?
Go to the Ensembl homepage and click on the link Tools at the top of the page. Currently there are nine tools listed in that page. Click on Variant Effect Predictor and enter the three variants as below:
7 117530985 117530985 G/A
7 117531038 117531038 T/C
7 117531068 117531068 T/C
Note: Variation data input can be done in a variety of formats. See more details about the different data formats and their structure in this VEP documentation page. Click Run. When your job is listed as Done, click View Results.
You will get a table with the consequence terms from the Sequence Ontology project (http://www.sequenceontology.org/) (i.e. synonymous, missense, downstream, intronic, 5’ UTR, 3’ UTR, etc) provided by VEP for the listed SNPs. You can also upload the VEP results as a track and view them on Location pages in Ensembl. SIFT and PolyPhen are available for missense SNPs only. For two of the entered positions, the variations have been predicted to have missense consequences of various pathogenicity (coordinate 117531038 and 117531068), both affecting CFTR. All the three variants have been already annotated and are known as rs1800077, rs1800078 and rs35516286 in dbSNP (databases, literature, etc).
VEP analysis of structural variants in human
We have details of a genomic deletion in a breast cancer sample in VCF format:
13 32307062 sv1 . <DEL> . . SVTYPE=DEL;END=32908738
Use VEP in Ensembl to find out the following information:
1. How many genes have been affected?
2. Does the structural variant (SV) cause deletion of any complete transcripts?
3. Map your variant in the Ensembl browser on the Region in detail view.
- Click on VEP at the top of any Ensembl page and open the web interface. Make sure your species is Human. It is good practise to name your VEP jobs something descriptive, such as Patient deletion exercise. Paste the variant in VCF format into the Paste data field and hit Run.
12 different genes are affected by the SV.
- Filter your table by select Consequence is
transcript_ablationat the top of the table and click Add.Yes, there is deletion of complete transcripts of PDS5B, N4BP2L1, BRCA2, RNY1P4, IFIT1P1, ATP8A2P2, N4BP2L2, N4BP2L2-IT2 and one gene without official symbols: ENSG00000212293.
- To view your variant in the browser click on the location link in the results table 13: 32307062-32908738. The link will open the Region in detail view in a new tab. If you have given your data a name it will appear automatically in red. If not, you may need to Configure this page and add it under the Personal data tab in the pop-up menu.







