Determining the effect of variants on genes with the VEP, demo
We have identified four variants on Anopheles gambiae chromosome 2R, an A deletion at 13225320, G->A at 14048788, T->C at 13550867 and G->A at 13952419.
We will use the Ensembl VEP to determine:
- Have my variants already been annotated in Ensembl?
- What genes are affected by my variants?
- Do any of my variants affect gene regulation?
Go to the front page of Ensembl Metazoa and click on _Tools _in the top bar.
Click on Variant Effect Predictor to open the input form.

Click on Add/remove species and find Anopheles gambiae.
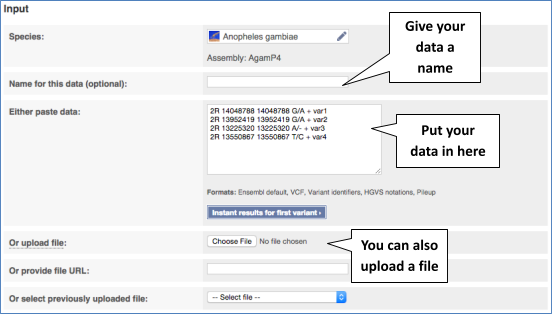
The data is in the format:
Chromosome Start End alleles (reference/alternative) strand name
Put the following into the Paste data box:
2R 14048788 14048788 G/A + var1
2R 13952419 13952419 G/A + var2
2R 13225320 13225320 A/- + var3
2R 13550867 13550867 T/C + var4
The VEP will automatically detect that the data is in Ensembl default format.
There are further options that you can choose for your output. These are categorised as Identifiers and frequency data, Filtering options and Extra options. Let’s open all the menus and take a look.
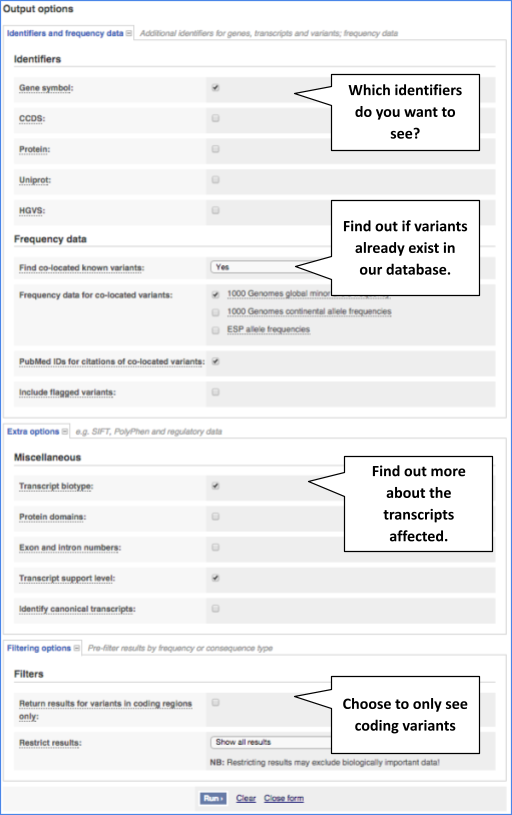
Hover over the options to see definitions.
When you’ve selected everything you need, scroll right to the bottom and click Run.
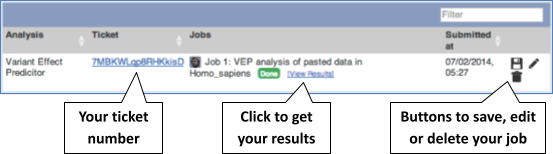
The display will show you the status of your job. It will say Queued, then automatically switch to Done when the job is done, you do not need to refresh the page. You can edit or discard your job at this time. If you have submitted multiple jobs, they will all appear here.
Click View results once your job is done.
In your results you will see a graphical summary of your data, as well as a table of your results. (Note that some empty columns in the results table have been hidden in the following screenshot to save space.)







