## Demonstration of the VEP web interface ### Input We have identified three variants on coffee chromosome 4: T -> C at 25759812 G -> A at 25685623 T -> G at 25863697 We will use the Ensembl VEP to determine which genes are affected by my variants? Click on **Tools** in the top green bar from any Ensembl Plants page, then **Variant Effect Predictor** to open the input form: 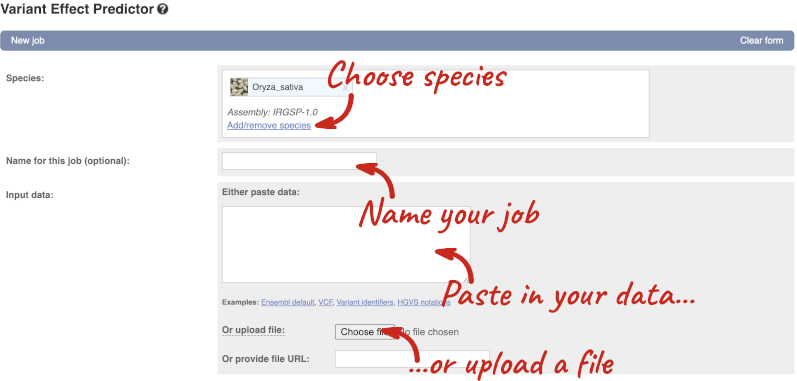 Click on **Add/remove species** and search for **_Triticum aestivum_** to choose it. The data is in VCF: `chromosome coordinate id reference alternative` Put the following into the Paste data box: ``` 4 25759812 var1 T C 4 25685623 var2 G A 4 25863697 var3 T G ``` The VEP will automatically detect that the data is in VCF. ### Additional configurations There are further options that you can choose for your output. These are categorised as **Identifiers, Variants and frequency data, Additional annotations, Predictions, Filtering options** and **Advanced options**. Let’s open all the menus and take a look. 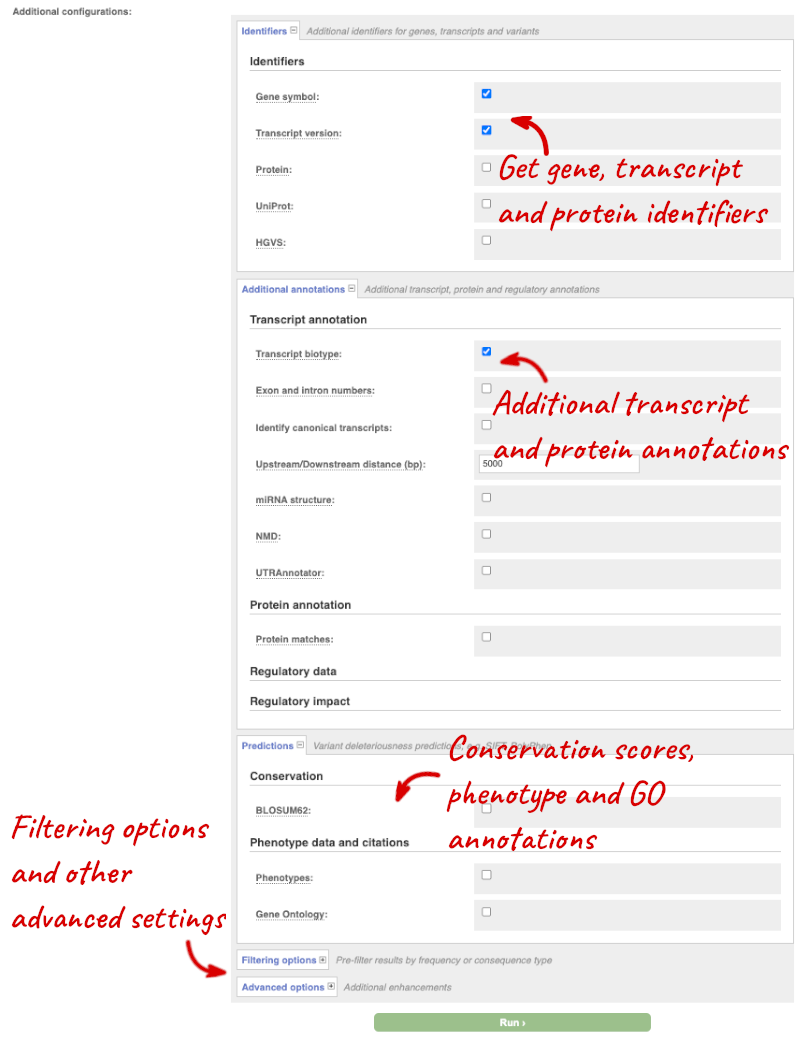 Hover over the options to see definitions. When you’ve selected everything you need, scroll right to the bottom and click **Run**.  The display will show you the status of your job. It will say **Queued**, then automatically switch to **Done** when the job is done, you do not need to refresh the page. You can edit or discard your job at this time. If you have submitted multiple jobs, they will all appear here. Click _View results_ once your job is done. ### Results In your results you will see a graphical summary of your data, as well as a table of your results. 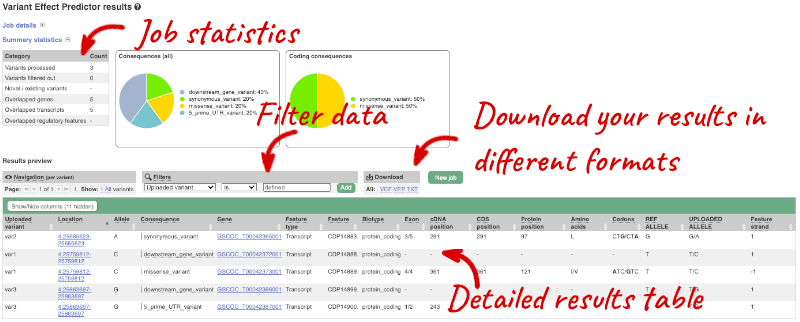 The results table is enormous and detailed, so we're going to go through the it by section. The first column is **Uploaded variant**. If your input data contains IDs, like ours does, the ID is listed here. If your input data is only loci, this column will contain the locus and alleles of the variant. You'll notice that the variants are not neccessarily in the order they were in in your input. You'll also see that there are multiple lines in the table for each variant, with each line representing one transcript or other feature the variant affects. You can mouse over any column name to get a definition of what is shown. The next few columns give the information about the feature the variant affects, including the consequence. Where the feature is a transcript, you will see the gene symbol and stable ID and the transcript stable ID and biotype. The IDs are links to take you to the gene or transcript homepage. 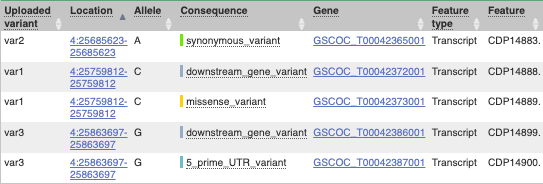 This is followed by details on the effects on transcripts, including the position of the variant in terms of the exon number, cDNA, CDS and protein, as well as the amino acid and codon change. 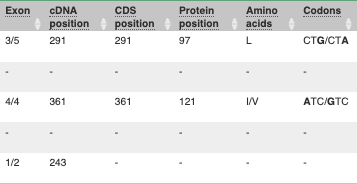 Above the table is the _Filter_ option, which allows you to filter by any column in the table. You can select a column from the drop-down, then a logic option from the next drop-down, then type in your filter to the following box. We'll try a filter of **Consequence**, followed by **is** then **missense_variant**, which will give us only variants that change the amino acid sequence of the protein. You'll notice that as you type `missense_variant`, the VEP will make suggestions for an autocomplete. 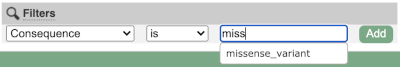 You can export your VEP results in various formats, including VCF. When you export as VCF, you'll get all the VEP annotation listed under `CSQ` in the `INFO` column. After filtering your data, you'll see that you have the option to export only the filtered data. You can also drop all the genes you've found into the Gene BioMart, or all the known variants into the Variation BioMart to export further information about them.