Filter Events by Year
Ensembl Variation – Multiplex assays of variant effects (MAVEs): Approaches, Analysis, and Interpretation
Course Details
- Lead Trainer
- Sarah Hunt
- Associate Trainer(s)
- Event Date
- 2025-11-26
- Location
- Hinxton, UK
- Description
- Work with the Ensembl Outreach team to get hands-on experience accessing and analysing variation data with the Ensembl genome browser.
Demos and exercises
VEP
We have identified five variants on human chromosome nine, C-> A at 128203516, an A deletion at 128328461, C->A at 128322349, C->G at 128323079 and G->A at 128322917.
We will use the Ensembl VEP to determine:
- Have my variants already been annotated in Ensembl?
- What genes are affected by my variants?
- Do any of my variants affect gene regulation?
Go to the front page of Ensembl and click on the Variant Effect Predictor.

This page contains information about the VEP, including links to download the script version of the tool. Click on Launch VEP to open the input form:
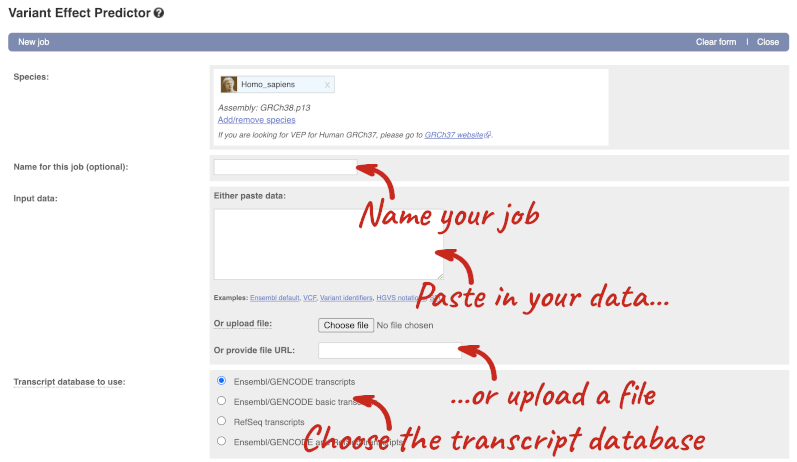
The data is in VCF format:
chromosome coordinate id reference alternative
Put the following into the Paste data box:
9 128328460 var1 TA T
9 128322349 var2 C A
9 128323079 var3 C G
9 128322917 var4 G A
9 128203516 var5 C A
The VEP will automatically detect that the data is in VCF.
There are further options that you can choose for your output. These are categorised as Identifiers, Variants and frequency data, Additional annotations, Predictions, Filtering options and Advanced options. Let’s open all the menus and take a look.
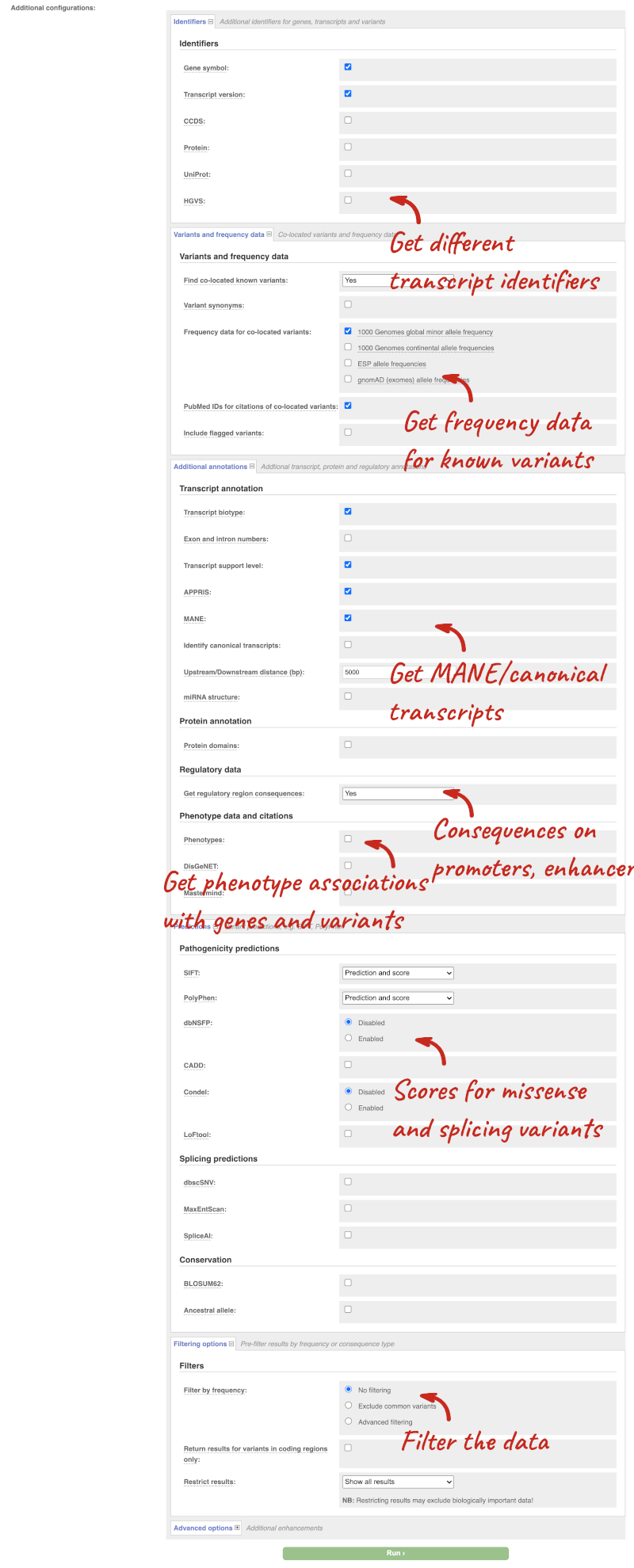
Hover over the options to see definitions.
We’re going to select some options:
- HGVS, annotation of variants in terms of the transcripts and proteins they affect, commonly-used by the clinical community
- Phenotypes
- Protein domains
When you’ve selected everything you need, scroll right to the bottom and click Run.

The display will show you the status of your job. It will say Queued, then automatically switch to Done when the job is done, you do not need to refresh the page. You can edit or discard your job at this time. If you have submitted multiple jobs, they will all appear here.
Click View results once your job is done.
In your results you will see a graphical summary of your data, as well as a table of your results.
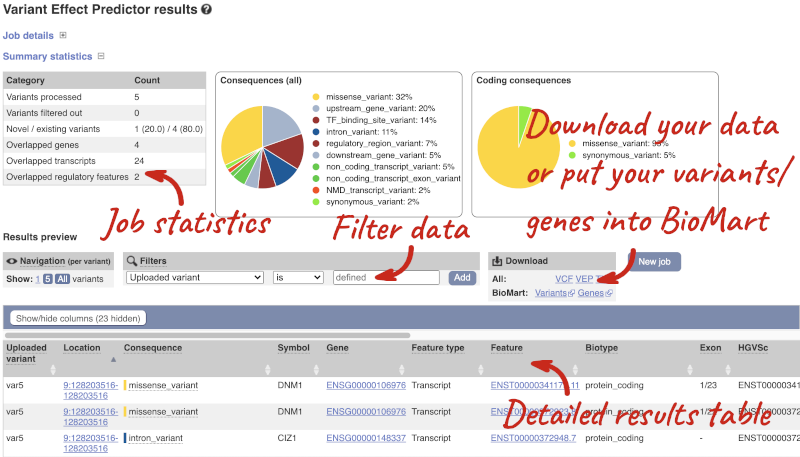
The results table is enormous and detailed, so we’re going to go through the it by section. The first column is Uploaded variant. If your input data contains IDs, like ours does, the ID is listed here. If your input data is only loci, this column will contain the locus and alleles of the variant. You’ll notice that the variants are not neccessarily in the order they were in in your input. You’ll also see that there are multiple lines in the table for each variant, with each line representing one transcript or other feature the variant affects.
You can mouse over any column name to get a definition of what is shown.
The next few columns give the information about the feature the variant affects, including the consequence. Where the feature is a transcript, you will see the gene symbol and stable ID and the transcript stable ID and biotype. Where the feature is a regulatory feature, you will get the stable ID and type. For a transcription factor binding motif (labelled as a MotifFeature) you will see just the ID. Most of the IDs are links to take you to the gene, transcript or regulatory feature homepage.
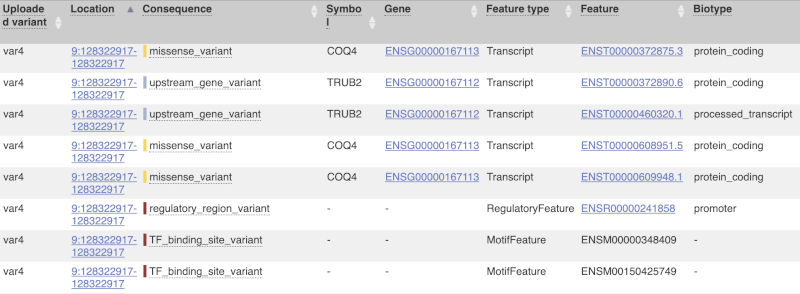
This is followed by details on the effects on transcripts, including the position of the variant in terms of the exon number, cDNA, CDS and protein, the amino acid and codon change, transcript flags, such as MANE, on the transcript, which can be used to choose a single transcript for variant reporting, and predicted pathogenicity scores. The predicted pathogenicity scores are shown as numbers with coloured highlights to indicate the prediction, and you can mouse-over the scores to get the prediction in words. Two options that we selected in the input form are the HGVS notation, which is shown to the left of the image below and can be used for reporting, and the Domains to the right. The Domains list the proteins domains found, and where there is available, provide a link to the 3D protein model which will launch a LiteMol viewer, highlighting the variant position.
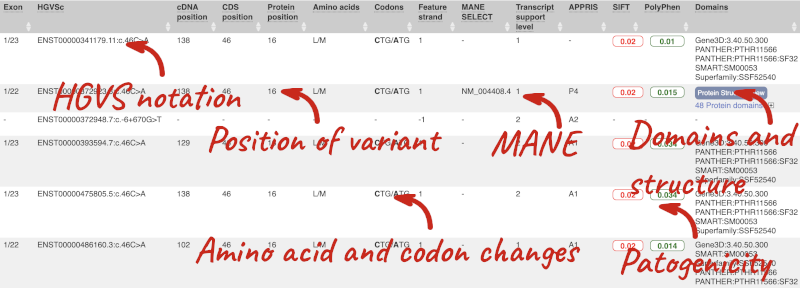
Where the variant is known, the ID of the existing variant is listed, with a link out to the variant homepage. In this example, only rsIDs from dbSNP are shown, but sometimes you will see IDs from other sources such as COSMIC. The VEP also looks up the variant from frequency files, and pulls back the allele frequency (AF in the table), which will give you the 1000 Genomes Global Allele Frequency. In our query, we have not selected allele frequencies from the continental 1000 Genomes populations or from gnomAD, but these could also be shown here. We can also see ClinVar clinical significance and the phenotypes associated with known variants or with the genes affected by the variants, with the variant ID listed for variant associations and the gene ID listed for gene associations, along with the source of the association.
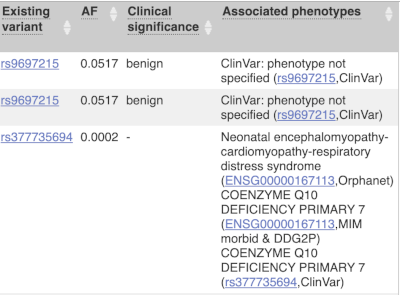
For variants that affect transcription factor binding motifs, there are columns that show the effect on motifs (you may need to click on Show/hide columns at the top left of the table to display these). Here you can see the position of the variant in the motif, if the change increases or decreases the binding affinity of the motif and the transcription factors that bind the motif.
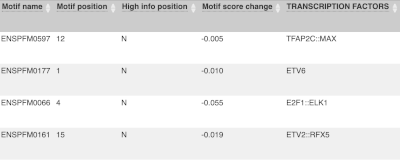
Above the table is the Filter option, which allows you to filter by any column in the table. You can select a column from the drop-down, then a logic option from the next drop-down, then type in your filter to the following box. We’ll try a filter of Consequence, followed by is then missense_variant, which will give us only variants that change the amino acid sequence of the protein. You’ll notice that as you type missense_variant, the VEP will make suggestions for an autocomplete.
You can export your VEP results in various formats, including VCF. When you export as VCF, you’ll get all the VEP annotation listed under CSQ in the INFO column. After filtering your data, you’ll see that you have the option to export only the filtered data. You can also drop all the genes you’ve found into the Gene BioMart, or all the known variants into the Variation BioMart to export further information about them.
Running CFTR variants through VEP
Resequencing of the genomic region of the human CFTR (cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) gene (ENSG00000001626) has revealed the following variants. The alleles defined in the forward strand:
- G/A at 7: 117,530,985
- T/C at 7: 117,531,038
- T/C at 7: 117,531,068
Use the VEP tool in Ensembl and choose the options to see SIFT and PolyPhen predictions. Do these variants result in a change in the proteins encoded by any of the Ensembl genes? Which gene? Have the variants already been found?
Go to the Ensembl homepage and click on the link Tools at the top of the page. Currently there are nine tools listed in that page. Click on Variant Effect Predictor and enter the three variants as below:
7 117530985 117530985 G/A
7 117531038 117531038 T/C
7 117531068 117531068 T/C
Note: Variation data input can be done in a variety of formats. See more details about the different data formats and their structure in this VEP documentation page. Click Run. When your job is listed as Done, click View Results.
You will get a table with the consequence terms from the Sequence Ontology project (http://www.sequenceontology.org/) (i.e. synonymous, missense, downstream, intronic, 5’ UTR, 3’ UTR, etc) provided by VEP for the listed SNPs. You can also upload the VEP results as a track and view them on Location pages in Ensembl. SIFT and PolyPhen are available for missense SNPs only. For two of the entered positions, the variations have been predicted to have missense consequences of various pathogenicity (coordinate 117531038 and 117531068), both affecting CFTR. All the three variants have been already annotated and are known as rs1800077, rs1800078 and rs35516286 in dbSNP (databases, literature, etc).
VEP analysis of structural variants in human
We have details of a genomic deletion in a breast cancer sample in VCF format:
13 32307062 sv1 . <DEL> . . SVTYPE=DEL;END=32908738
Use VEP in Ensembl to find out the following information:
1. How many genes have been affected?
2. Does the structural variant (SV) cause deletion of any complete transcripts?
3. Map your variant in the Ensembl browser on the Region in detail view.
- Click on VEP at the top of any Ensembl page and open the web interface. Make sure your species is Human. It is good practise to name your VEP jobs something descriptive, such as Patient deletion exercise. Paste the variant in VCF format into the Paste data field and hit Run.
12 different genes are affected by the SV.
- Filter your table by select Consequence is
transcript_ablationat the top of the table and click Add.Yes, there is deletion of complete transcripts of PDS5B, N4BP2L1, BRCA2, RNY1P4, IFIT1P1, ATP8A2P2, N4BP2L2, N4BP2L2-IT2 and one gene without official symbols: ENSG00000212293.
- To view your variant in the browser click on the location link in the results table 13: 32307062-32908738. The link will open the Region in detail view in a new tab. If you have given your data a name it will appear automatically in red. If not, you may need to Configure this page and add it under the Personal data tab in the pop-up menu.
Ensembl VEP - BAP1 MaveDB
Web VEP analysis of variants from a MaveDB dataset
Introduction
The Ensembl Variant Effect Predictor (VEP) is a powerful tool for annotating genomic variants. Ensembl VEP is accessible via web, REST API and command-line options.
In this practical session, we will practice using Ensembl VEP via the web interface to annotate variants in HGVS format. The variants we will use were from an assay of exon 10 in the Homo sapiens BAP1 gene from the MaveMD collection on MaveDB. You can access the data on MaveDB here via the Scores file.
Ensembl VEP tutorials
A general Ensembl VEP tutorial for the web interface is available on the EMBL-EBI Training website to try out or compare to if you need some guidance. You can use the Ensembl VEP web documentation pages to help find and understand the configurations you will need.
Launch the Ensembl VEP web interface via the Ensembl navigation bar or by clicking here.
Exercises
Ex 1. We will make use of variants from the Scores file mentioned above. Download the Scores file for BAP1 exon 10 from MaveDB, and open it in any tool that can view csv files. Note the “hgvs_nt” and the “scores” columns, as we will use these for input and filtering respectively.
Ex 2. Explore the page info at BAP1 exon 10 from MaveDB.
- What does the distribution of scores look like?
- Using the “Clinical view”, do you note any pattern of Pathogenic or benign variants across score bins?
- What specific transcript ID was used for this assay?
Ex 3. Explore that transcript information by searching it on the Ensembl website. What type of transcript is this? The transcript is flagged as the MANE Select transcript of the BAP1 gene. What does the MANE Select flag tell you?
Ex 4. Use the Ensembl VEP web interface to annotate the BAP1 exon 10 variants from Exercise 1. You will need to supply the list of HGVS format variants, either by extracting that column and uploading as a file, or by copy and pasting in the list of variants. Select the following Ensembl VEP options and any others that interest you:
- enable “gnomAD (genomes) allele frequencies”
- enable “Protein Matches”
- enable “AlphaMissense” pathogenicity prediction
- enable “SpliceAI” splicing predictions
- make sure “Gene Symbol” is enabled
- make sure “MANE” is enabled
When you have enabled those options, submit the job with “Run”.
Ex 5. Once the job completes, explore your Ensembl VEP results on the web interface to answer the below. Download your results in TXT format so you can count the number of results.
- How many of these variants have been observed in the population?
- What is the most common coding consequence?
- How many results are in the output text file, why are there more than the number you submitted?
Ex 6. Use the Ensembl VEP filter options to filter your results for only those corresponding to the feature (transcript ID) to that identified in Exercise 2. Export the filtered TXT file.
Ex 7. Using the scores file from MaveDB, extract the variants with with scores lower than -0.09 only. Use this list to filter your Ensembl VEP result and explore the consequence types of these, are there any patterns that you notice? You may use the awk and grep bash commands for this step to help you, or you can try other ways of merging the input table with your Ensembl VEP results.
Exercise 1
Open the file “urn_mavedb_00000662-k-1_scores.csv” in any text editor or by using a command such as less -S urn_mavedb_00000662-k-1_scores.csv
If you have a csv viewer like libreoffice, googledocs, excel, you can open up this comma delimited file to view. Hint, you can use a command like awk -F',' '{print $2}' urn_mavedb_00000662-k-1_scores.csv > bap1_mdb_hgvs.txt to extract the IDs from column 2 to use as input for Ensembl VEP. Mind to remove the header when pasting/uploading file. In case of server issues, the file used for this practical is available here.
Exercise 2
Most scores are close to 0, in bins ranging between -0.02 and 0.02. The Clinical view shows there are a few pathogenic variants around the score range of 0.10. This assay targeted the Coding Sequence (CDS), splice-site containing intron and 3’UTR of the transcript: ENST00000460680.6.
Exercise 3
It is a protein coding transcript. MANE select transcripts are those where Ensembl/Gencode (ENST) and RefSeq (NM) transcripts that are 100% identical (5’UTR, CDS and 3’UTR) and 3) are highly conserved, expressed and well-supported.
Exercise 4
Your Ensembl VEP job should take a few mins to run. If you find a blank result, check that you have only included the hgvs and no header or other info as input. In case of server issues, a pre-run output file is available here
Exercise 5
The summary table on the top of the Ensembl VEP results show Novel / existing variants 656 (57.1) / 493 (42.9). Missense variant is the most common coding consequence. There are many more result lines than input variants, as Ensembl VEP will report consequences for all transcripts and genes overlapped by the variant coordinate.
19794 lines in vep_bap1_result.txt
Exercise 6
In the filter options, use the drop down menu to select “Feature” “is” and paste ENST00000460680.6 then click “Add”. This limits the result to only those in the same transcript used in the assay. You may export the file, and in case of server issues, the transcript filter file is here
Exercise 7
It’s possible to programming languages and tools to merge the tables, but for this practical, we will use awk and grep to extract to simplify for this exercise. When working at high volumes of variants, we recommend automating steps.
First use awk to filter the table for scores lower than -0.09:
awk -F',' 'NR==1; $3<-0.09' urn_mavedb_00000662-k-1_scores.csv> bap1_exon10_low_scores.txt
That command keeps the header line intact. Run another awk command to extract only the variant names “hgvs_nt”:
awk -F',' '{print $2}' bap1_exon10_low_scores.txt > bap1_exon10_hgvs_low_scores.txt
You can use that file as input into grep to filter the Ensembl VEP results from Exercise 6:
grep -f bap1_exon10_hgvs_low_scores.txt vep_bap1_ex10_maveDB.Feature_is_ENST00000460680.6.txt > vepbap1_low_scores.txt
The consequence type is primarly made up of stop gained and frameshift variants. Do these align with the scores measured by the assay given the assay type? Are any variant types here unexpected?
NB - if you have access to a command line version of Ensembl VEP, this practical can use a command such as: ./vep --af --af_gnomadg --appris --biotype --buffer_size 500 --check_existing --distance 5000 --domains --mane --polyphen b --pubmed --regulatory --show_ref_allele --sift b --species homo_sapiens --symbol --transcript_version --tsl --uploaded_allele --cache --input_file [input_data] --output_file [output_file]
Splice AI and AlphaMissense are plugins so you would need to download and install these to retrieve scores.







