Filter Events by Year
Ensembl Bacteria – Microbial Genomics, Decode Life
Course Details
- Lead Trainer
- Louisse Paola Mirabueno
- Associate Trainer
- Event Dates
- 2025-02-03 until 2025-02-04
- Location
- Virtual
- Description
- We will explore Ensembl Bacteria to learn more about the latest data and where it came from, view annotation of molecular interactions involving bacterial genes and comparative genomics data involving key species
- Survey
- Ensembl Bacteria – Microbial Genomics, Decode Life Feedback Survey
Demos and exercises
Species and genome assemblies
Demo: Introduction to Ensembl
Ensembl
Homepage
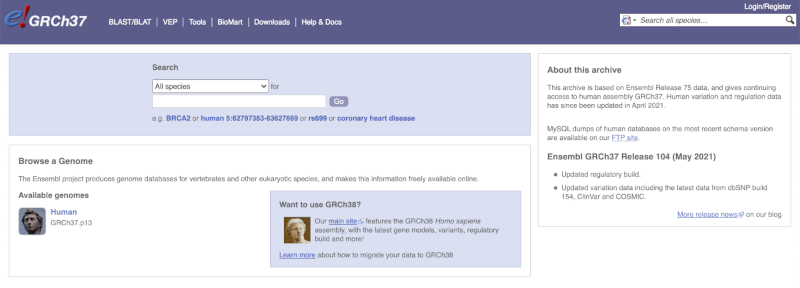
The front page of Ensembl is found at ensembl.org. It contains lots of information and links to help you navigate Ensembl:
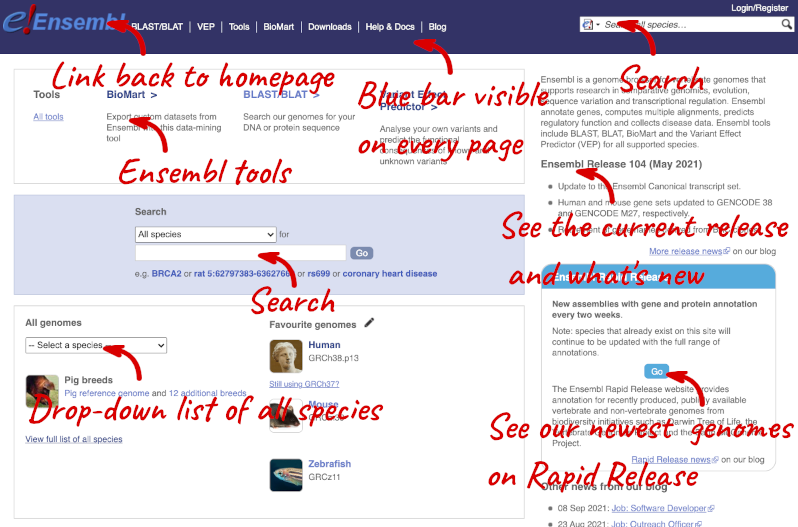
On the right-hand panel you can see the current release number and what has come out in this release. To access old releases, scroll to the bottom of the page and click on View in archive site in the right-hand corner.
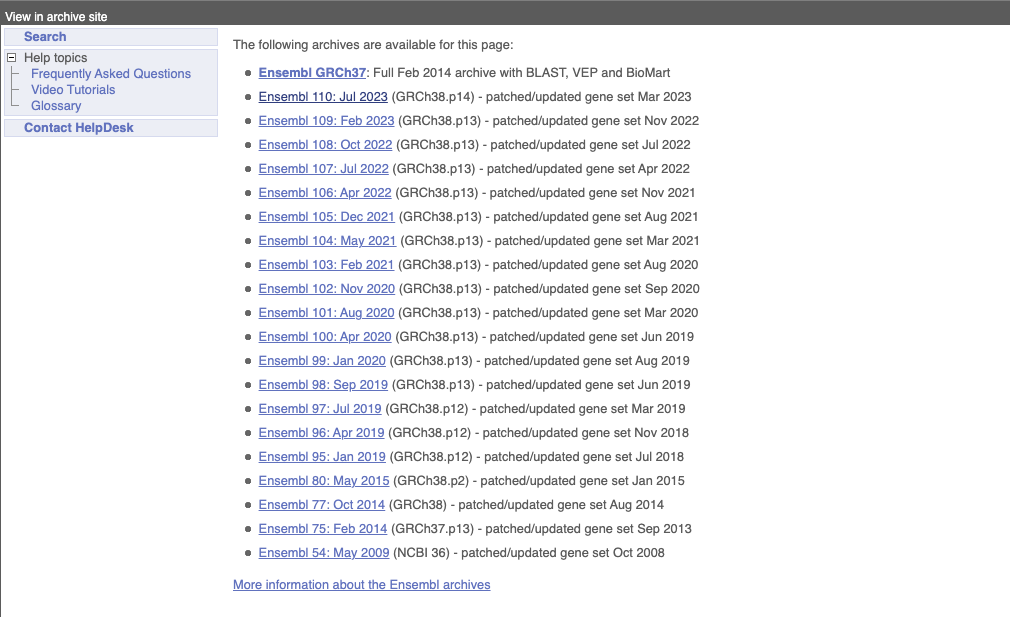
Click on the links to go to the archives. Alternatively, you can jump quickly to the correct release by adding e plus the release number in the URL. For example e98.ensembl.org jumps to Ensembl release 98.
Available species
Scroll back up to the top of the homepage. You can view all available species by clicking the View full list of all species link underneath the coloured search block.
You can search for your species of interest (either the common or scientific name) using the search bar at the top right-hand corner of the table. Click on the common name of your species of interest to go to the species information page. We’ll click on Human.
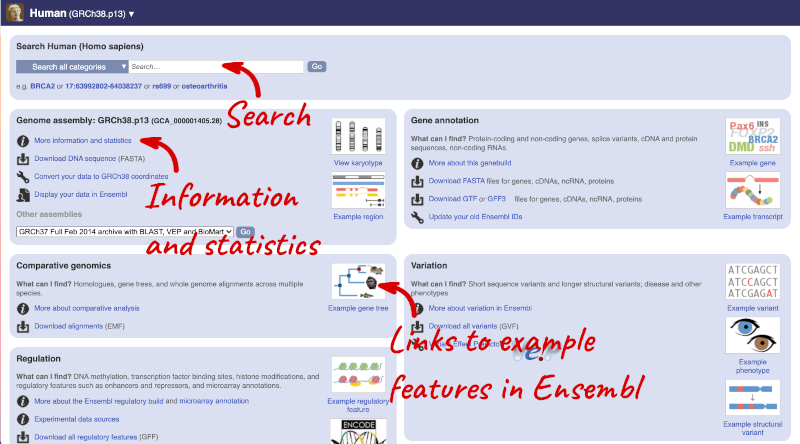
Species information
Here you can see links to example features and to download flatfiles. To find out more about the genome assembly and genebuild, click on More information and statistics under the Genome assembly section.
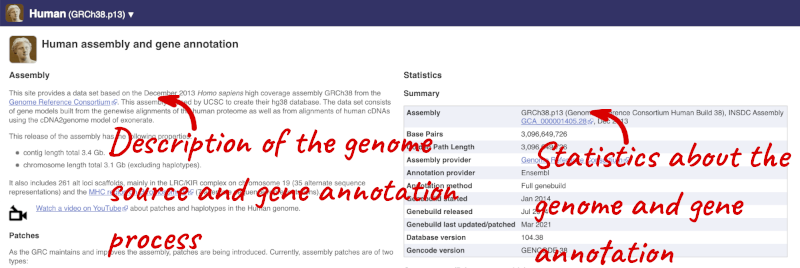
Here you’ll find a detailed description of how to the genome was produced and links to the original source. You will also see details of how the genes were annotated.
The current genome assembly for human is GRCh38. If you want to see the previous assembly, GRCh37, visit our dedicated site grch37.ensembl.org.
Ensembl Genomes
Homepage
Let’s take a look at the Ensembl Genomes homepage at ensemblgenomes.org.
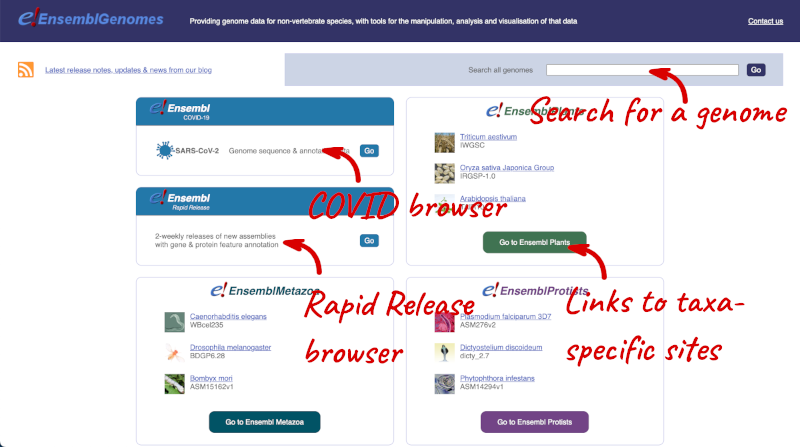
Click on the different taxa to see their homepages. Each one has a different colour-coding, but they are all structured in a similar format to the Ensembl main site.
You can navigate most of the taxa in the same way as you would with Ensembl.
Ensembl Bacteria
Ensembl Bacteria has a large number of genomes and has a slightly different method to the other Ensembl sites. Let’s look at it in more detail.
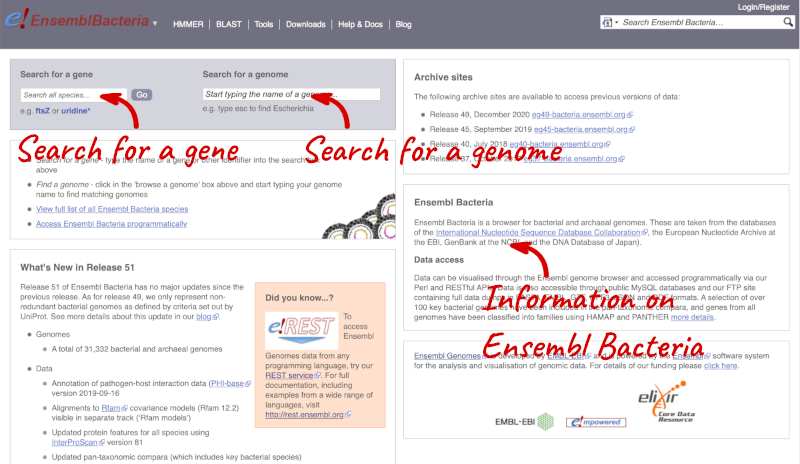
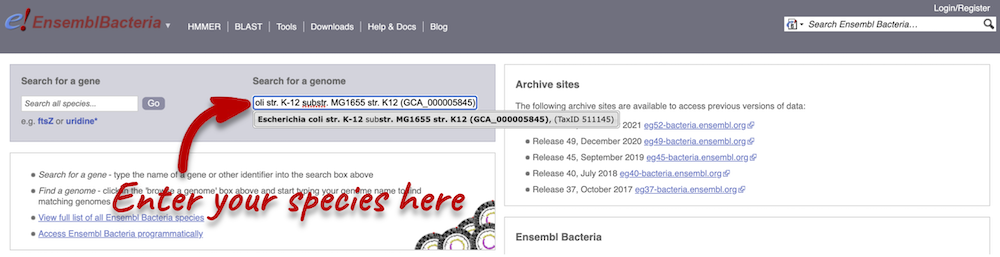
There’s no drop-down species list for bacteria as it would be hard to navigate with the number of species. You can click the View full list of all Ensembl Bacteria species link underneath the coloured search block. Search for your species of interest using the filter in the top right-hand corner of the table.
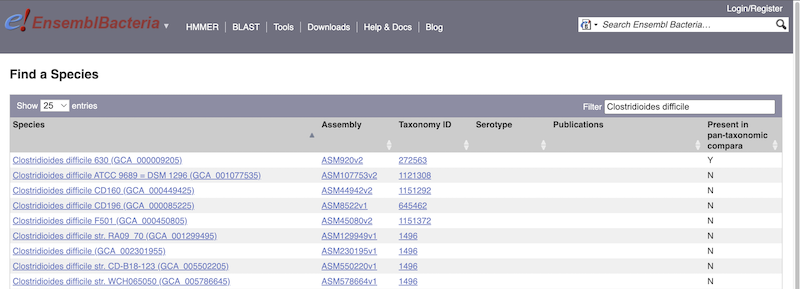
Alternatively, you can find a species by typing the species name into the Search for a genome search box at the top of the page. A drop-down list will appear with any species matching the name you entered.
For example, to find a sub-strain of Clostridioides difficile start typing in the species name. Due to the auto-complete, you’ll see useful results as soon as you get to Clostridio.
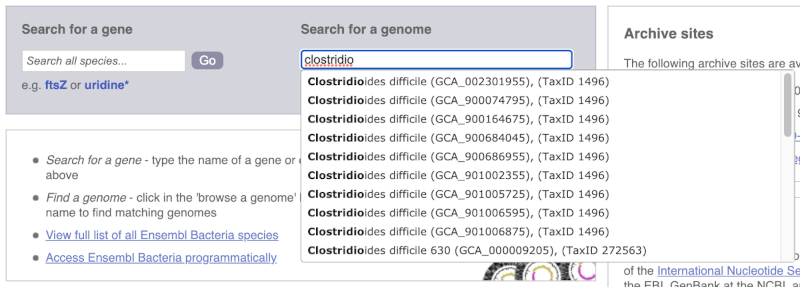
The drop down contains various strains of C. difficile. Let’s choose C. difficile 630. This will take us to another species information page, where we can explore various features.
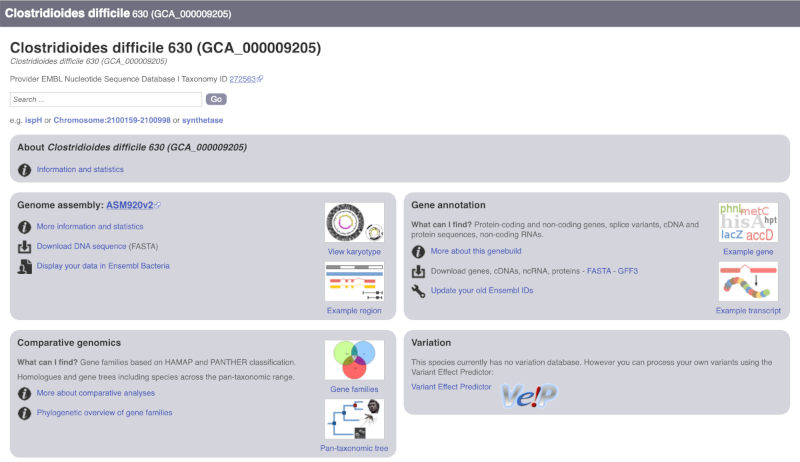
Unlike the Homo sapiens species information page, there is no prose description of the genome or gene annotation, as these pages were generated automatically.
Ensembl Rapid Release
Our newest genomes, such as those coming from the Darwin Tree of Life, are available rapid.ensembl.org with limited annotation.
Finding a genome in Ensembl Bacteria
Mycobacterium tuberculosis H37Ra str. ATCC25177 is a clinical strain.
Go to Ensembl Bacteria and find the species M. tuberculosis H37Ra str. ATCC25177. How many coding genes does it have?
In the Ensesmbl Bacteria homepage, start to type H37Ra into the Search for a genome search box (you can find this in the coloured block at the top of the homepage). It will auto-complete, allowing you to select M. tuberculosis H37Ra str. ATCC25177 from the drop-down list. Click on More information and statistics.
M. tuberculosis H37Ra str. ATCC25177 has 4,080 coding and 47 non-coding genes.
Exploring genomic regions
Region in Detail view
Start at the Ensembl Bacteria homepage, bacteria.ensembl.org. Search for your species of interest either by using the search box, or opening the full list of species by clicking View full list of all Ensembl Bacteria species underneath the search box.
Enter Escherichia coli str. K-12 substr. MG1655 (GCA_000005845) in the search box. Enter Chromosome:3144663-3157453 into the species-specific search box:
Press Enter or click Go to jump directly to the Region in detail page.
Click on the  button to view page-specific help. The help pages provide text, labelled images and, in some cases, help videos to describe what you can see on the page and how to interact with it.
button to view page-specific help. The help pages provide text, labelled images and, in some cases, help videos to describe what you can see on the page and how to interact with it.
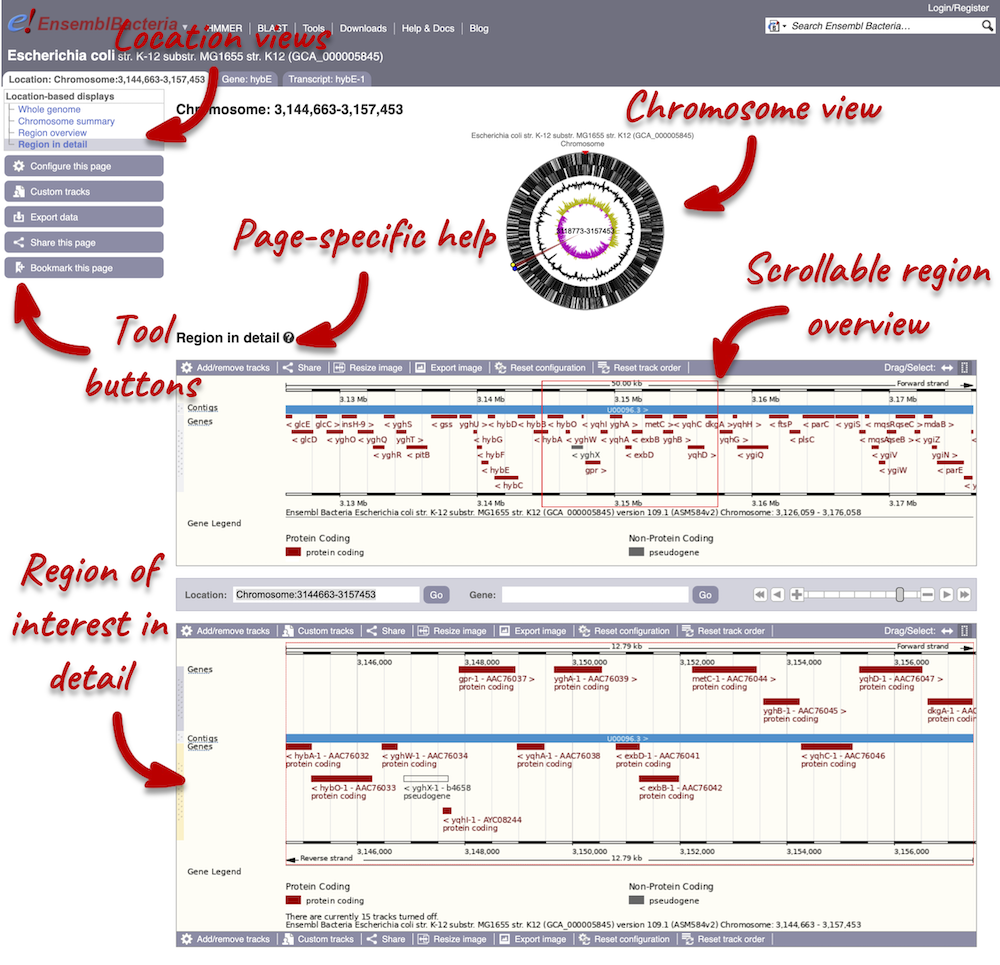
The Region in detail page is made up of three images, let’s look at each one on detail.
The first image shows the chromosome:
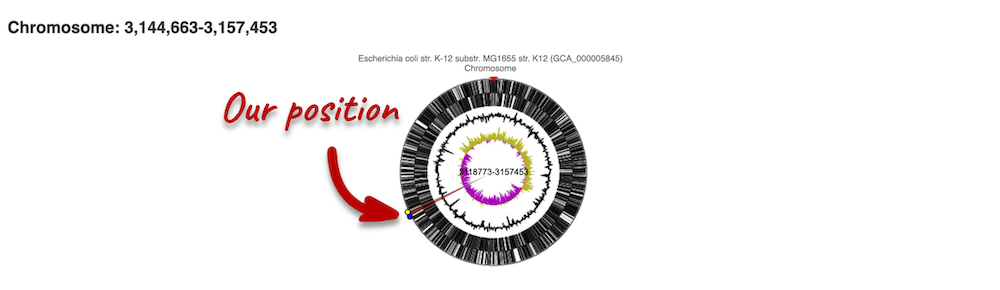
You can jump to a different region by clicking and dragging the yellow and blue handles.
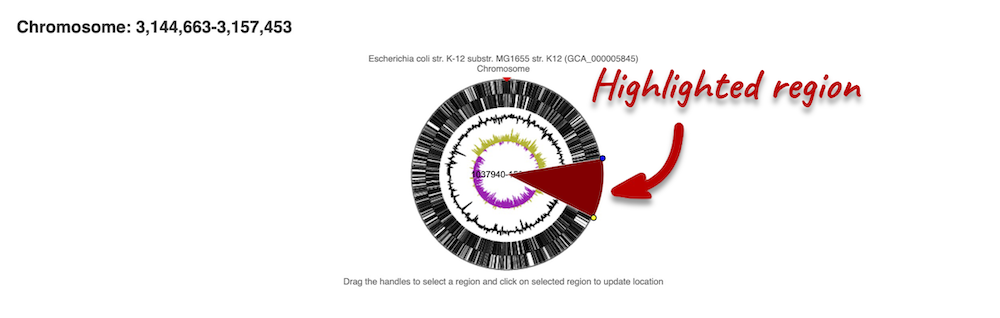
If you want to move to your highlighted region, you click on the region shaded in red.
The second image shows a 50 kb region (the size varies per genome and depends on the gene size and density; you can find a scale at the top of the view) around our selected region. This view allows you to scroll back and forth along the chromosome.
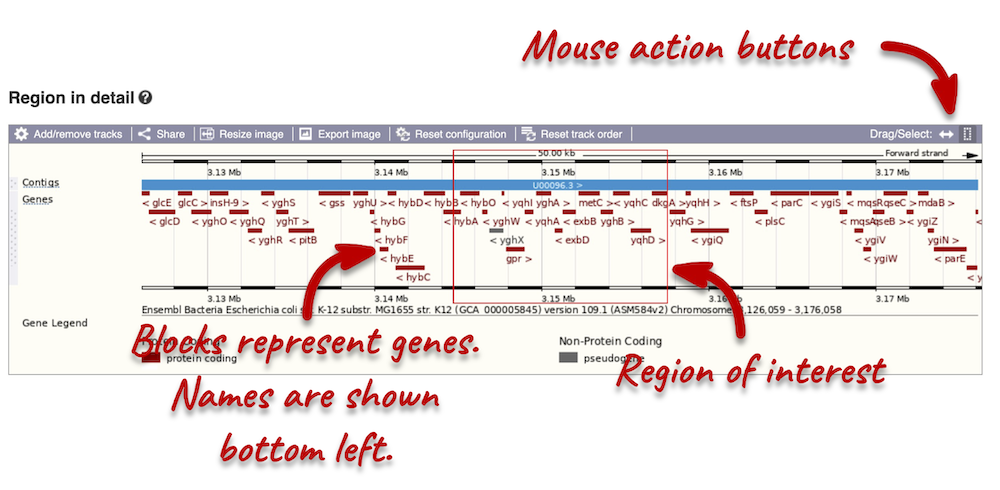
Click and drag your mouse to highlight a region. A pop-up window will appear with options to jump to or centre on the highlighted region.
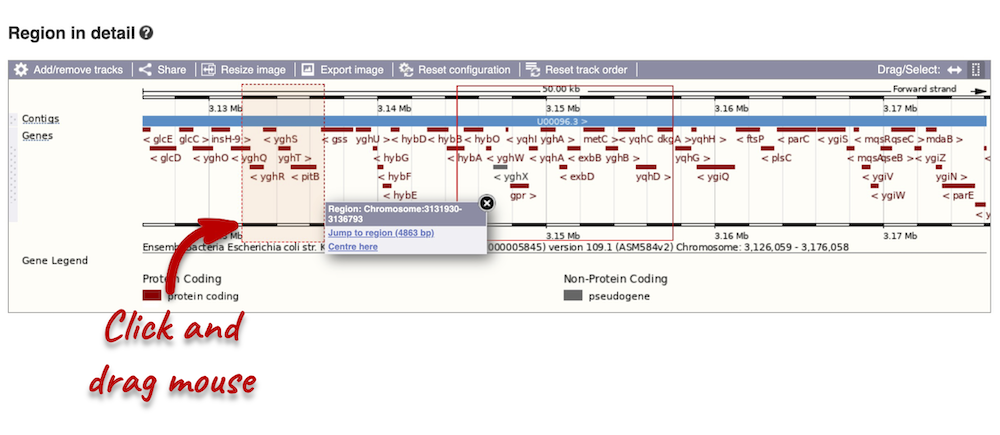
Click on the X to close the pop-up menu.
Click on the Drag/Select button  to change the action of your mouse click. Now you can scroll along the chromosome by clicking and dragging within the image. As you do this you’ll see the image below grey out and update to your scrolled region. To go back to go back to where you started, you can click the Back button of your browser.
to change the action of your mouse click. Now you can scroll along the chromosome by clicking and dragging within the image. As you do this you’ll see the image below grey out and update to your scrolled region. To go back to go back to where you started, you can click the Back button of your browser.
The third image is a detailed, configurable view of the region.
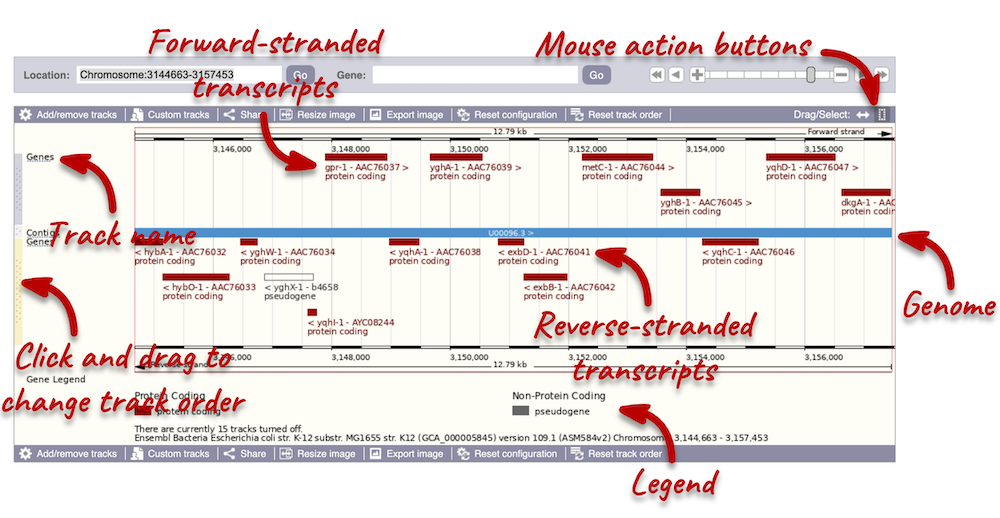
Click on the Drag/Select option at the top or bottom right to switch mouse action. On Drag, you can click and drag left or right to move along the genome, the page will reload when you drop the mouse button. On Select you can drag out a box to highlight or zoom in on a region of interest.
We can edit what we see on this page by clicking on the blue Configure this page menu at the left.

This will open a menu that allows you to change the image.
You can enable tracks in different styles; more details are in the FAQs. 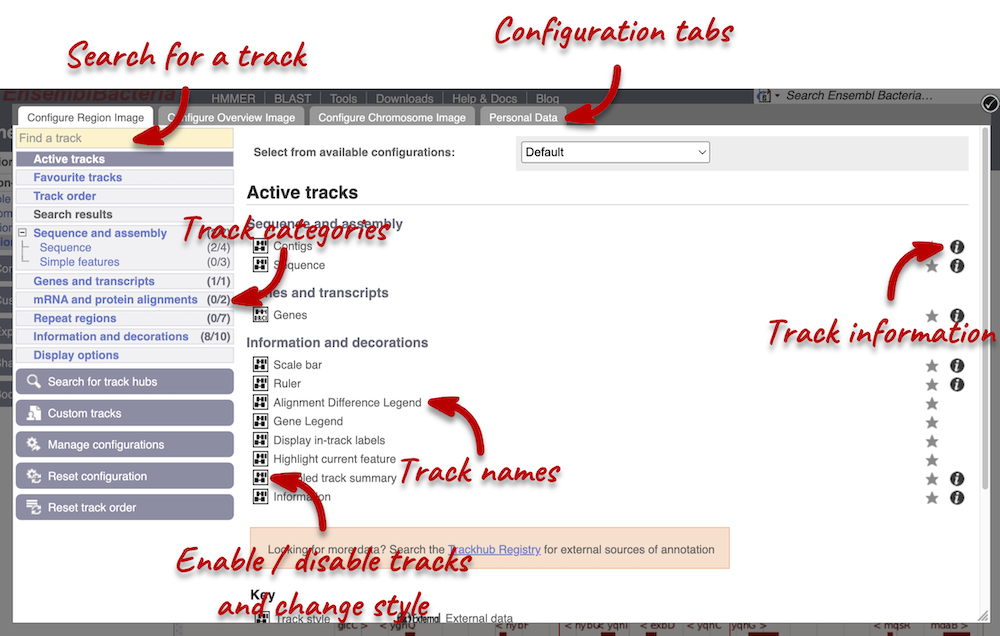
Let’s add the following tracks to our view:
- Start/stop codons
- All repeats
Now click on the check icon in the top left-hand corner to save and close the menu. Alternatively, click anywhere outside of the menu. We can now see the tracks in the image.
We can also change the way the tracks appear by clicking on the track name then hovering over the cog wheel to open its menu. We can move tracks around by clicking and dragging on the bar to the left of the track name.
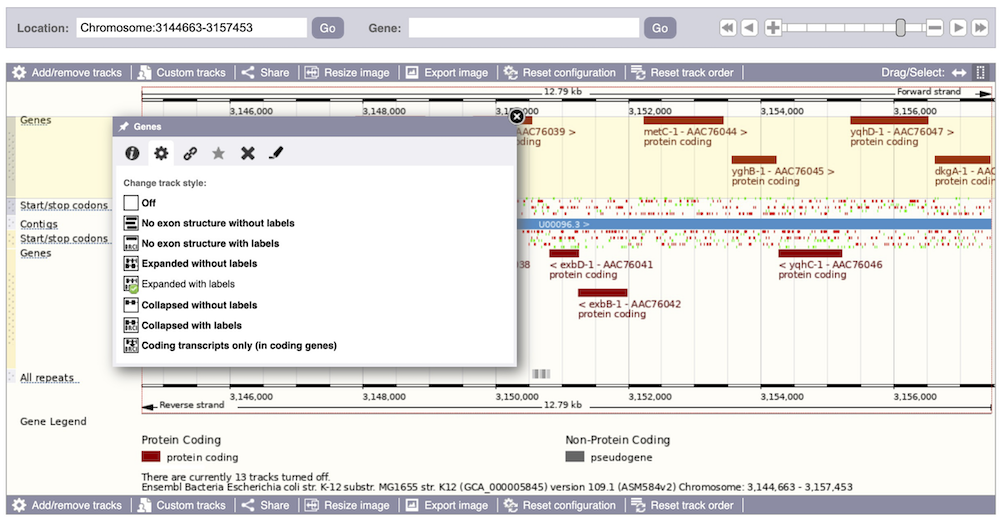
Now that you’ve got the view how you want it, you might like to show something you’ve found to a colleague or collaborator. Click on the Share this page button to generate a URL with your set configurations. Email the link to someone else, so that they can see the same view as you, including all the tracks you’ve added. These links contain the Ensembl release number, so if a new release or even assembly comes out, your link will just take you to the archive site for the release it was made on.
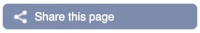
To return this to the default view, go to Configure this page and select Reset configuration at the bottom of the menu.
Exploring a genomic region in Staphylococcus aureus
Go to the Ensembl Bacteria homepage and do the following:
-
Search for the Staphylococcus aureus subsp. aureus NCTC 8325 (GCA_000013425).
-
Search for the gene gyrA.
-
What are the genomic coordinates of this gene? Is gyrA located on the forward or reverse strand?
-
Name two genes located upstream and downstream of gyrA.
-
On the Ensembl bacteria homepage, type
NCTC 8325into the Search for a genome box. Click on the auto-completed genome name to navigate to the species information page. -
Type gyrA into the search box. Click Go.
-
The coordinates of the gyrA are 7,005-9,668. The gene is located on the forward strand.
-
SAOUHSC_00005 (DNA gyrase, B subunit) is located upstream gyrA, and SAOUHSC_00007 (a conserved hypothetical protein) is located downstream gyrA.
Exploring a genomic region in Salmonella enterica
Go to Ensembl Bacteria and do the following:
-
Search for the Salmonella enterica subsp. enterica serovar Typhi str. Ty2 (GCA_000007545) (Hint: type
Tyinto the Search for a genome box). -
Go to the region Chromosome:2000605-2009742.
-
How many genes are annotated in this region? How many are on the forward strand? How many are on the reverse strand?
-
Go to the Ensembl Bacteria homepage. Type
Ty2into the Search for a genome box. Click on the auto-completed genome name to navigate to the species information page. -
Type
Chromosome:2000605-2009742into the search box. Click Go. -
There are 8 genes annotated in this region, all on the reverse strand.
Gene annotation
Demo: Viewing genes and transcripts
You can find out lots of information about Ensembl genes and transcripts using the browser. If you’re already looking at a Region in detail view, you can click on any transcript and a pop-up menu will appear, allowing you to jump directly to that gene or transcript.
Alternatively, you can find a gene by searching for it. You can search for gene names, identifiers, or functions that might be associated with the genes.
We’re going to look at the lacZ gene Escherichia coli str. K-12 substr. MG1655 (GCA_000005845). From bacteria.ensembl.org, search for the Escherichia coli_ str. K-12 substr. MG1655 (GCA_000005845) genome. Type lacZ into the species-specific search bar and click the Go button.
The gene tab
Click on the gene ID b0344 from the search hits. The Gene tab should open:
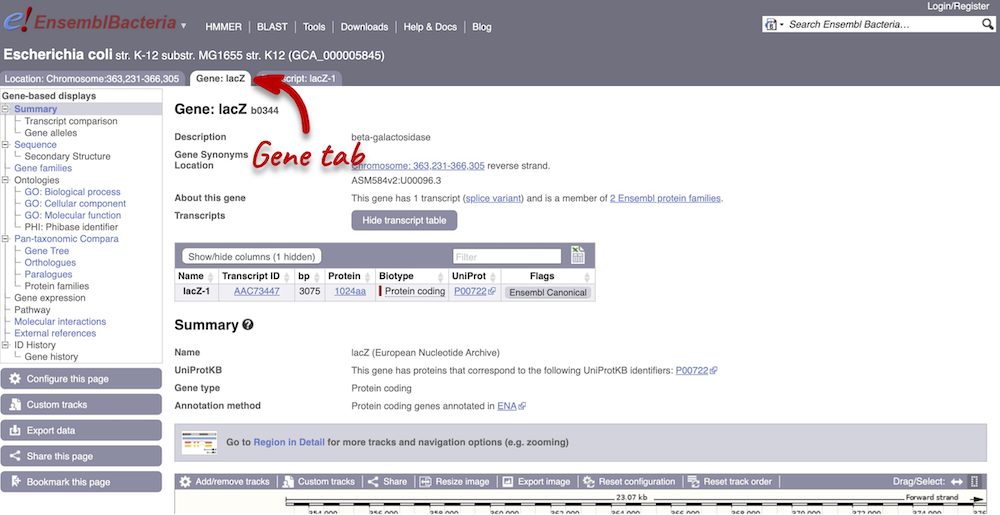
This page summarises the gene, including its location, name and equivalents in other databases. At the bottom of the page, a graphic shows a Region in detail view with the transcripts. We can also see the overlapping and neighbouring genes.
There are different tabs for different types of features, such as genes and transcripts. These appear side-by-side underneath the species name at the top of the page, allowing you to jump back and forth between features of interest. Each tab has its own navigation column down the left hand-side of the page, listing all the things you can see for this feature.
Gene sequence
Let’s walk through the menu for the Gene tab. Click Sequence in the left-hand panel to view the genomic sequence of the gene.
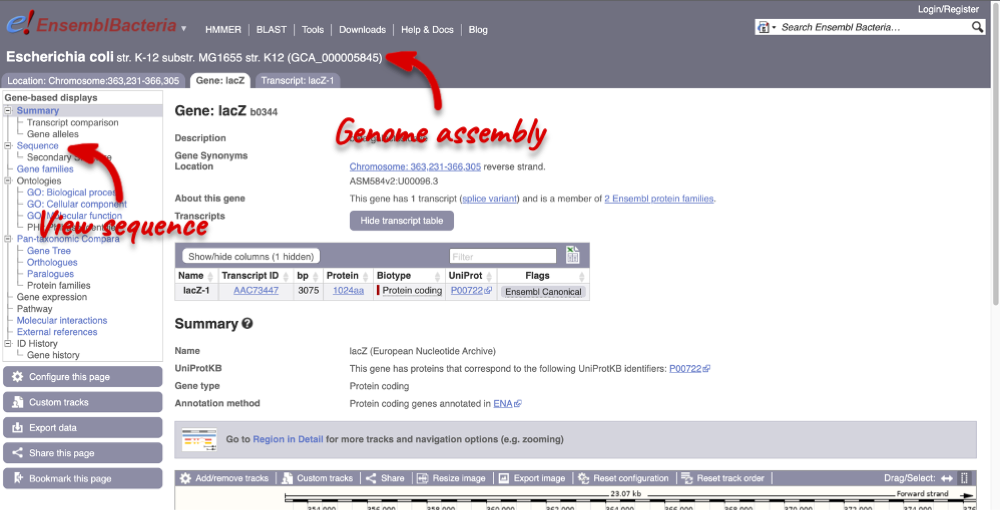
The sequence is shown in FASTA format. The FASTA header contains the genome assembly, chromosome, coordinates and strand (1 or -1). This gene is on the positive strand.

Exons are highlighted within the genomic sequence: the exon of our gene of interest and any neighbouring or overlapping genes. By default, 600 bases are shown up and downstream of the gene. We can make changes to how this sequence appears with the Configure this page button found at the left. This allows us to change the flanking regions, add line numbering and more. Click on it now.
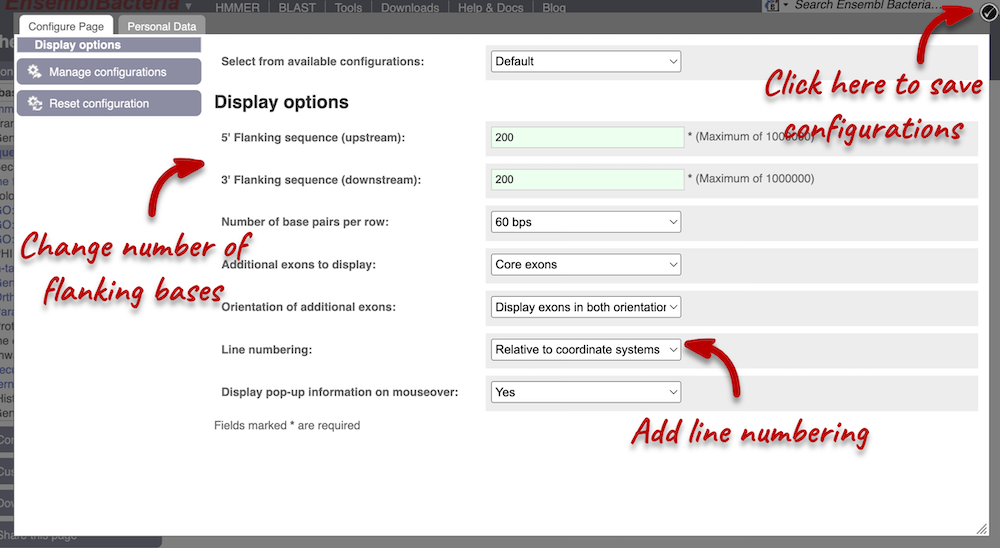
We have changed our Flanking sequences to 200 and added Line numbering relative to the coordinate system. Save your setting by clicking the check icon at the top right-hand corner.
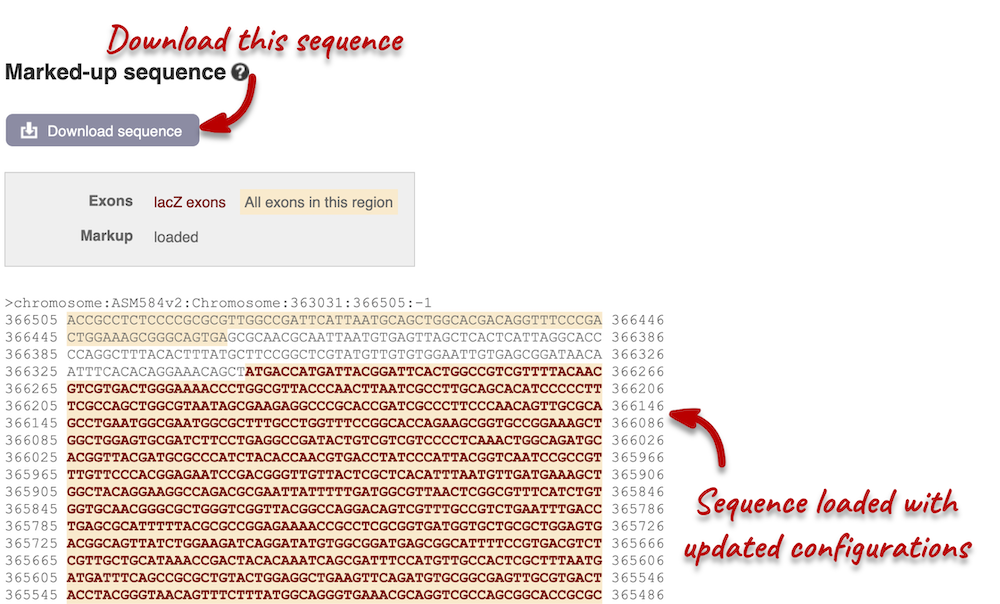
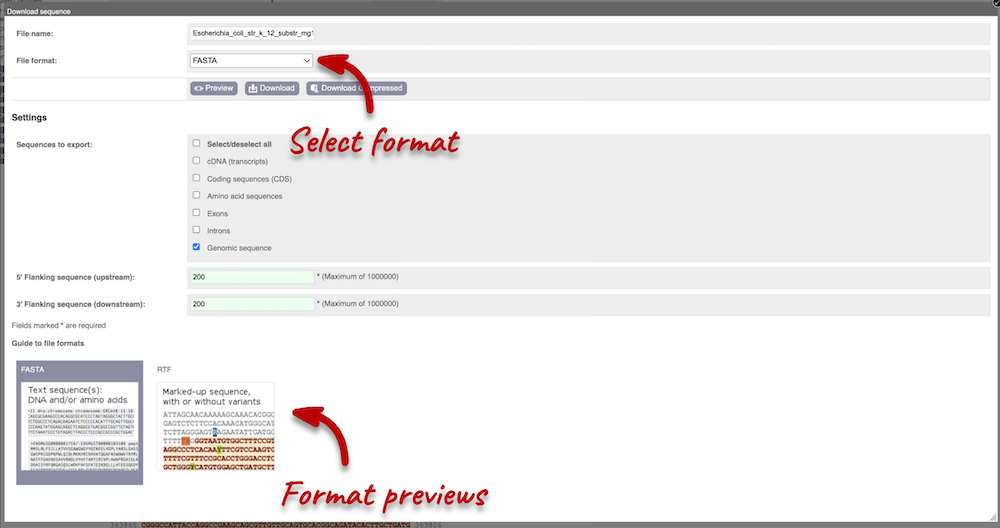
You can download this sequence by clicking in the Download sequence button above the sequence. This will open a dialogue box that allows you to pick between plain FASTA sequence, or sequence in rich-text format (RTF), which includes all the coloured annotations and can be opened in a word processor. If you want run a sequence analysis tool, download as FASTA sequence, whereas if you want to analyse the sequence visually, RTF is best for this. This button is available for all sequence views.
Gene function
To find out the protein function, have a look at gene ontology (GO) terms from the Gene Ontology consortium. There are three pages of GO terms, representing the three divisions: GO: Biological process (what the protein does)
GO: Cellular component (where the protein is)
GO: Molecular function (how it does it)
Click on GO: Biological process to see an example of the GO pages.
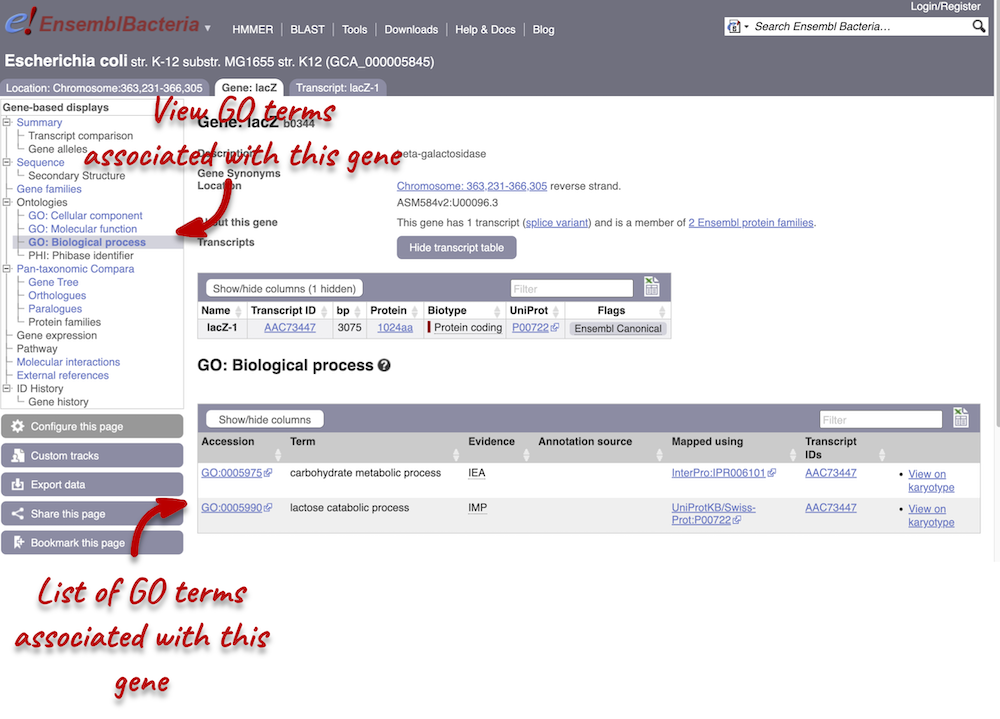
Here you can see the functions that have been associated with the gene. There are three-letter codes that indicate how the association was made, as well as links to the specific transcript they are linked to.
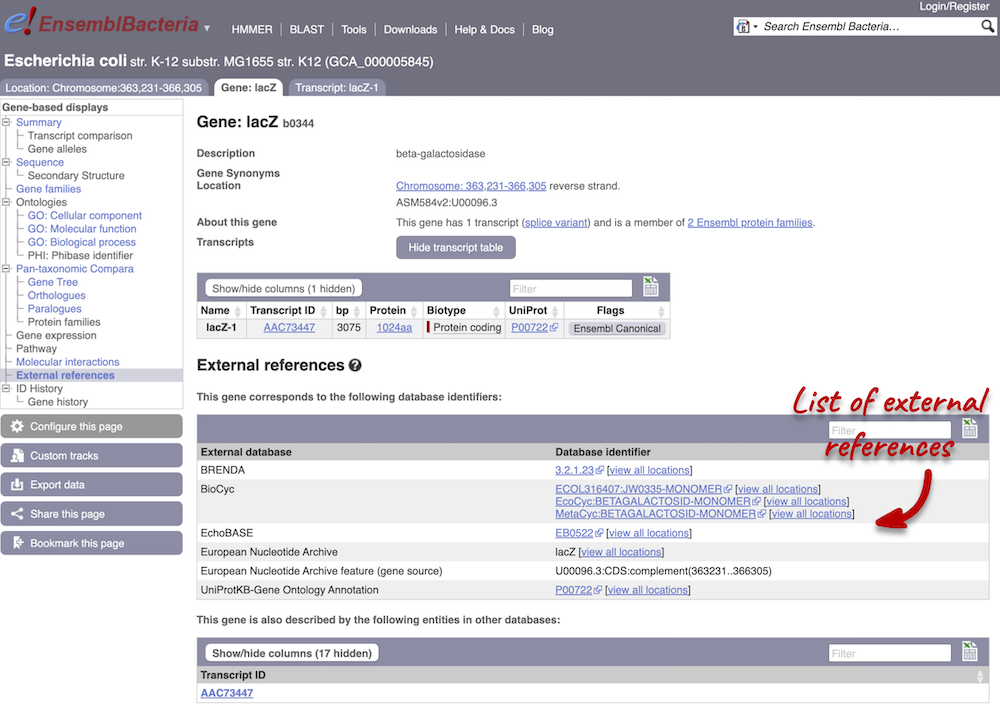
Gene information in external databases
We also have links out to other databases which have information about our genes and may focus on other topics that we don’t cover, like the European Nucleotide Archive ENA) or the UniProt knowledge base UniProtKB. Go up the left-hand menu to External references:
The transcript tab
We’re now going to explore the transcript of lacZ. Click on Show transcript table underneath the gene summary at the top of the page.

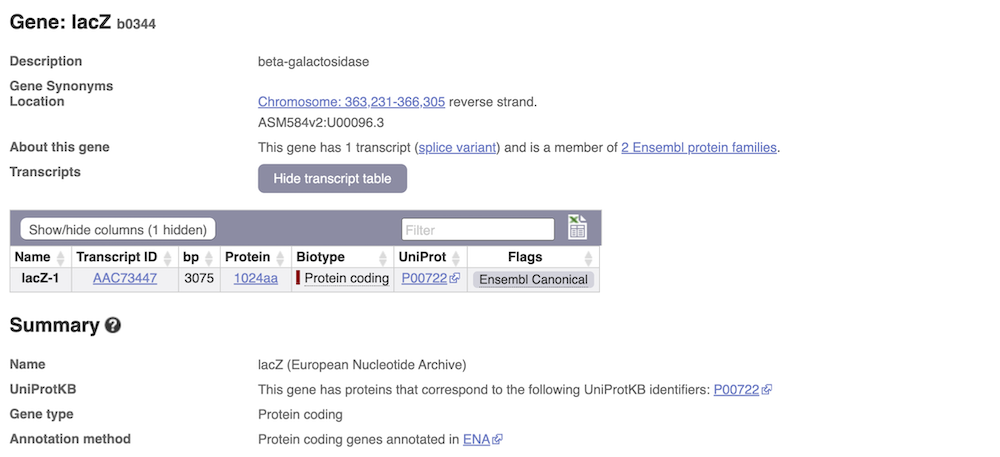
Here we can see a list of all the transcripts of lacZ with their identifiers, lengths and biotypes. The lacZ gene only has one transcript. Click on the transcript ID AAC73447.
You are now in the Transcript tab for AAC73447. We can still see the gene tab so we can easily jump back. The left hand navigation column provides several options for the transcript AAC73447 - many of these are similar to the options you see in the gene tab, but not all of them. If you can’t find the thing you’re looking for, often the solution is to switch tabs.
Transcript sequences
Click on the Exons link in the left-hand panel. This page is useful as it will give you the length of the coding sequence.
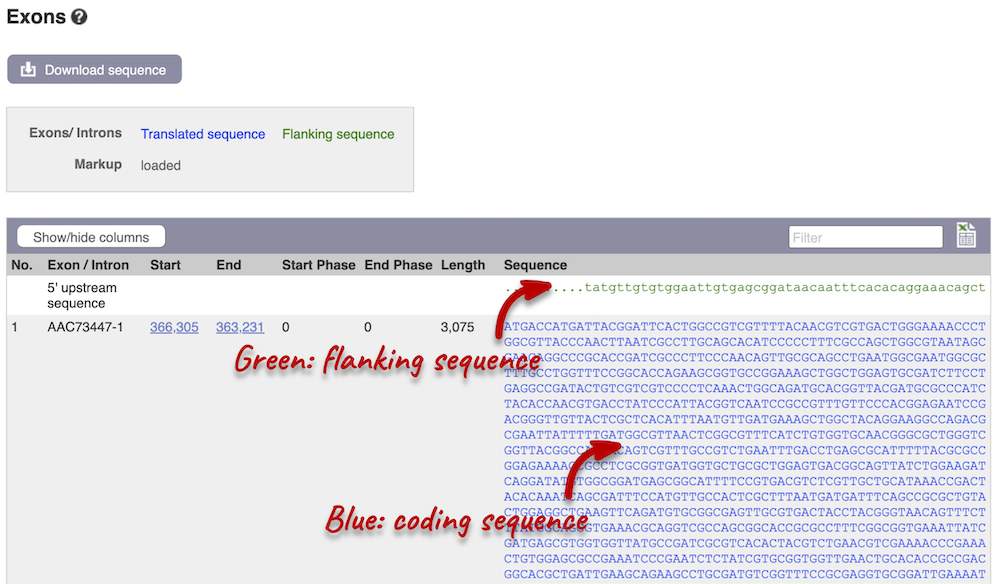
You may want to change the display (for example, to show more flanking sequences). In order to do so, click on Configure this page and change the display options accordingly.
Transcript information in external databases
Next, follow the General identifiers link at the left. Just like the External References page in the Gene tab, this page shows links out to other databases such as InterPro, PDB, UniProtKB, and others, this time linked to the transcript or protein product, rather than the gene.
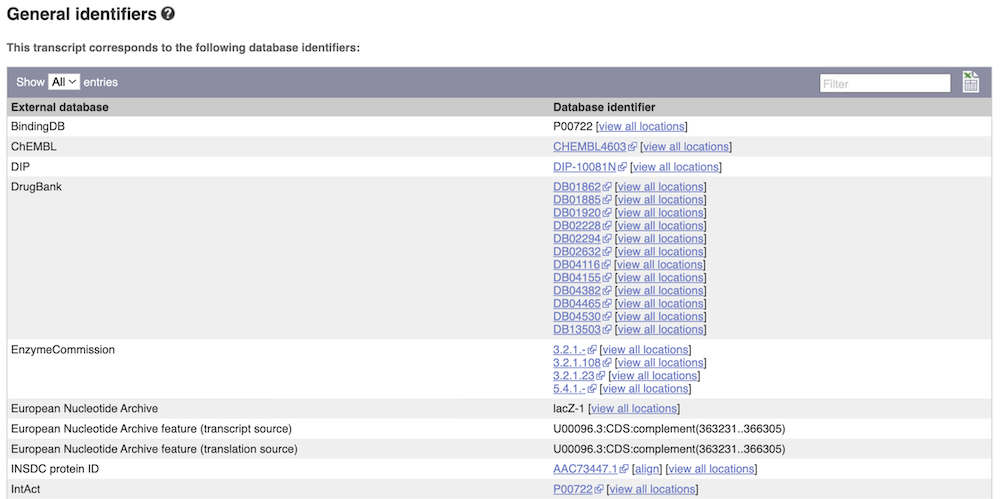
Protein domain information
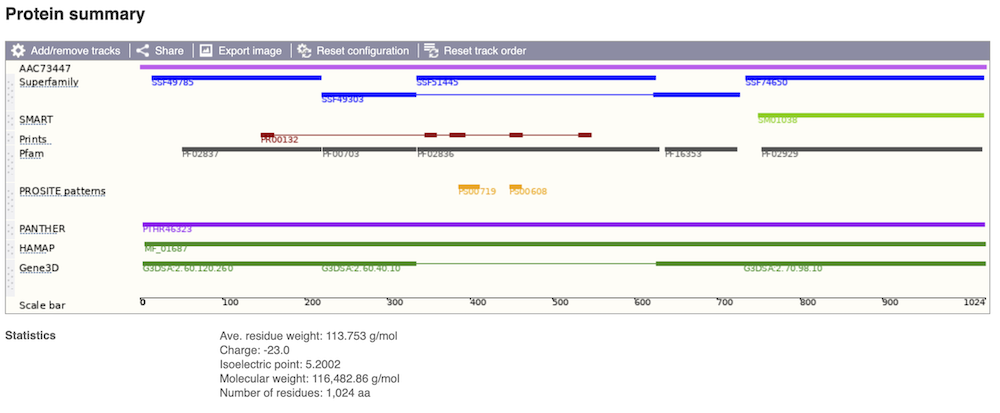
If you’re interested in protein domains, you could click on Protein summary to view domains from different sources, such as SMART and PROSITE. These are all plotted against the transcript sequence.
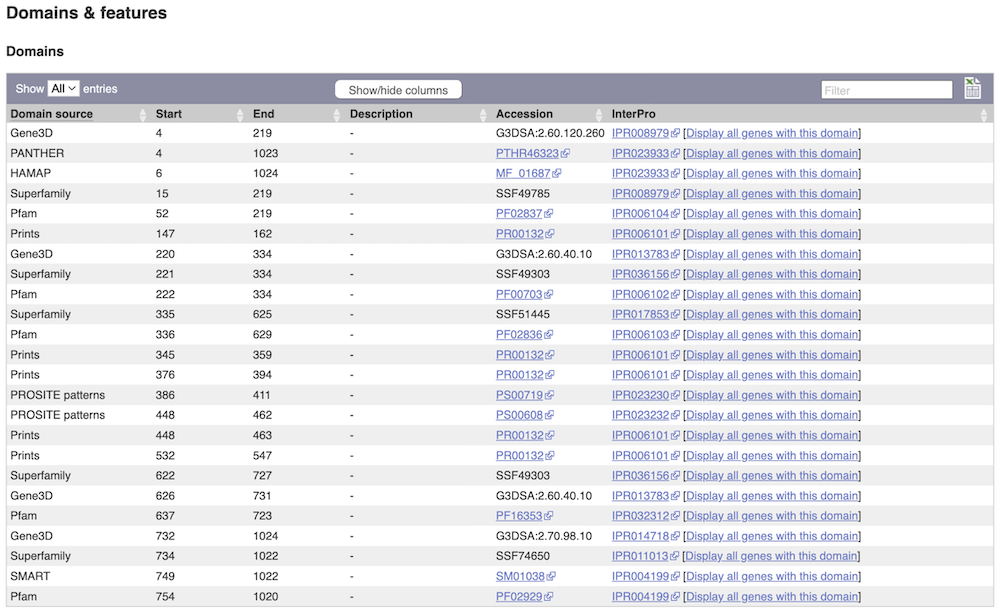
Alternatively, you can go to Domains & features to see a table of the same information in a tabular format.
Exploring a bacterial gene in Clostridium sporogenes
Start in Ensembl Bacteria and select the Clostridium sporogenes (GCA_001444695) genome.
-
What is the gene name for the Glutamine synthetase gene?
-
Go to the transcript tab. How long is the transcript? How long is the protein?
-
What domains can be found in the protein product of this transcript? How many different domain prediction methods agree with each of these domains?
- From the Ensembl Bacteria homepage, select Clostridium sporogenes by beginning to write the species name and selecting the species from the auto-complete list. Type
Glutamine synthetaseand click on the gene ID ENSB:yZtlLO8Ti90y75J which will open the Summary display on the Gene tab..The gene name is
glnA. - Switch to the Transcript tab and go to the Summary display. You can find the length under Statistics underneath the transcript image.
The
glnAtranscript is 1,899 bp and the protein is 632 aa in length. - Click on either Protein Summary or Domains & features in the left hand menu to see graphically or as a table respectively.
6 protein domains were found. All of them predict a glutamine synthetase domain.
Exploring a gene in Escherichia coli
Start in Ensembl Bacteria and search for the Escherichia coli str. K-12 substr. MG1655 (GCA_000005845) genome.
-
What GO: biological process terms are associated with the Era gene?
-
How many different InterPro domains are found in the protein product of this gene?
-
What is the associated UniProt ID of the transcript?
Enter part of the name into the genome search box (e.g. MG1655) and then select the correct genome to go to the species information page.
- Enter
Erainto the search box and hit Go. Click the link in the first hit to go to the era gene page. From here, click GO: Biological process in the left-hand menu.There are three GO IDs: GO:0000028, GO:0006468, GO:0042274 and GO:0046777.
- Switch to the Transcript tab and go to Domains & features in the left-hand panel. Count the number of unique InterPro IDs in the table.
8 different InterPro domains are found in the protein product of Era.
- You can find the UniProt ID in the transcript table or under General identifiers in the left-hand panel.
The UniProtKB/Swiss-Prot ID is P06616.
Ensembl VEP
Demo: VEP
Input
We have identified 5 variants on Saccharomyces cerevisiae R64-1-1 chromosome VII:
T -> C at 3598
G -> C at 3929
T -> G at 5566
T -> A at 5727
A -> T at 7628
We will use Ensembl VEP to determine:
- Have my variants already been annotated in Ensembl?
- What genes are affected by my variants?
- Are any of my variants missense variants?
Click on Tools in the navigation bar at the top of any Ensembl Fungi page, then click Variant Effect Predictor to open the input form:
Click on Add/remove species and search for Saccharomyces cerevisiae R64-1-1 to select it.
First, we need to convert our variants into one of the formats supported by VEP. You can find a list of input formats and examples in the VEP documentation page. Let’s put our data in VCF:
chromosome coordinate id reference alternative
Copy and paste the following data into the Input data box:
VII 3598 var1 T C
VII 3929 var2 G C
VII 5566 var3 T G
VII 5727 var4 T A
VII 7628 var5 A T
VEP will automatically detect that the data is in VCF.
Additional configurations
There are further options that you can choose for your output. These are categorised as Identifiers, Variants and frequency data, Additional annotations, Predictions, Filtering options and Advanced options. You can open all tabs to explore the different options.
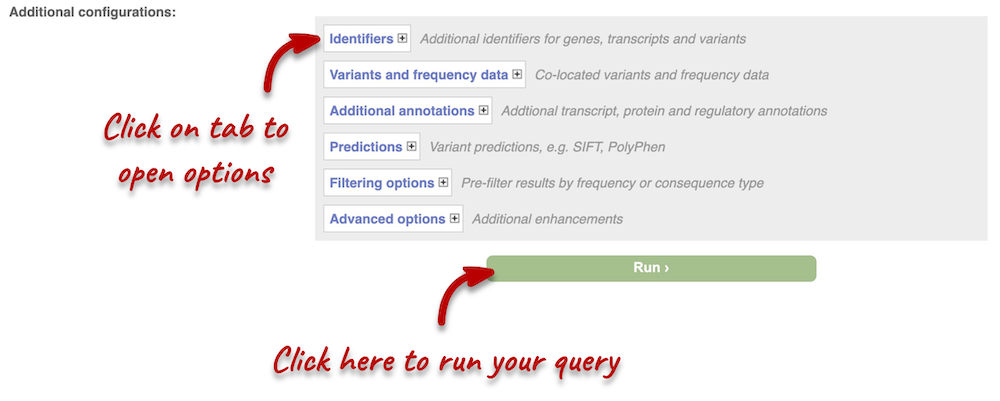
Hover over the options to see definitions. When you’ve selected everything you need, scroll right to the bottom and click Run.

The display will show you the status of your job. It will say Queued, then automatically switch to Done when the job is done, you do not need to refresh the page. You can edit or discard your job at this time. If you have submitted multiple jobs, they will all appear here.
Click View results once your job is done.
Results
In your results you will see a graphical summary of your data, as well as a table of your results.
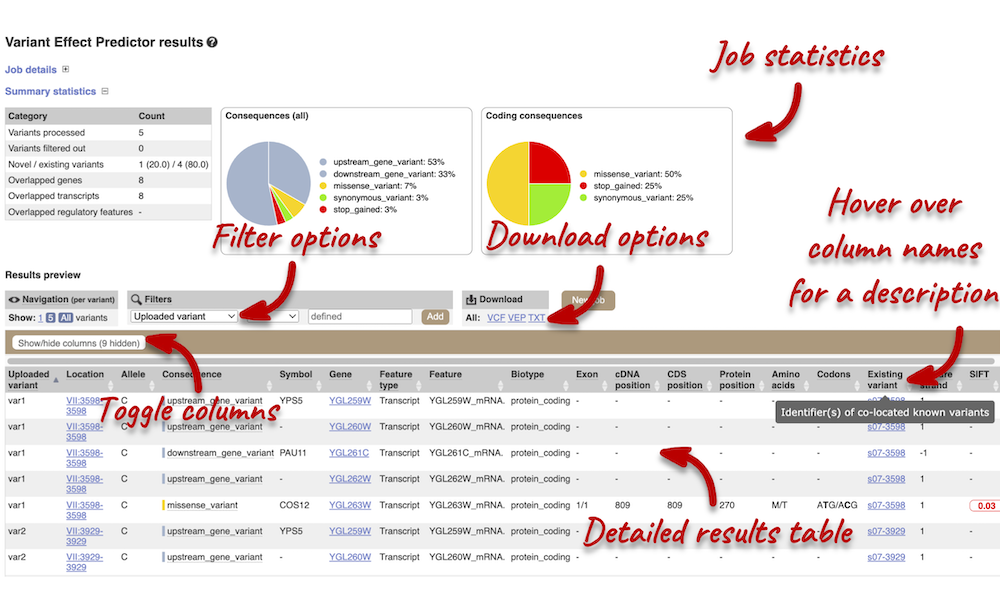
The results table is very detailed, so we’re going to go through the it by section. The first column is Uploaded variant. If your input data contains IDs, like ours does (i.e. var1, var2, etc.), the ID is listed here. If your input data is only loci, this column will contain the locus and alleles of the variant. You’ll notice that the variants are not neccessarily in the order they were in in your input. You’ll also see that there are multiple lines in the table for each variant, with each line representing one transcript or other feature the variant affects.
You can hover over any column name with your mouse to get a definition of what is shown.
The next few columns show information about the feature the variant affects, including the consequence. Where the feature is a transcript, you will see the gene symbol and stable ID and the transcript stable ID and biotype. The IDs are links to take you to the Gene or Transcript tabs.
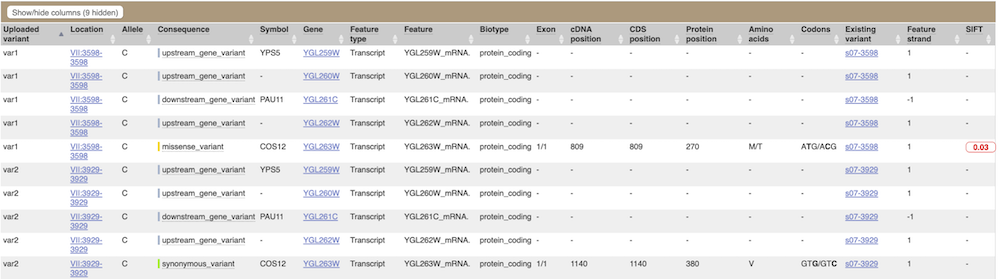
This is followed by details on the effects on transcripts, including the position of the variant in terms of the exon number, cDNA, CDS and protein, the amino acid and codon change and pathogenicity scores. Where the variant is known, the ID of the existing variant is listed under the column Existing variant, with a link out to the Variant tab. The pathogenicity scores are shown as numbers with coloured highlights to indicate the prediction, and you can mouse-over the scores to get the prediction in words.
Above the table is the Filter option, which allows you to filter by any column in the table. You can select a column from the drop-down, then a logic option from the next drop-down, then type in your filter to the following box. We’ll try a filter of Consequence, followed by is then missense_variant, which will give us only variants that change the amino acid sequence of the protein. You’ll notice that as you type missense_variant, the VEP will make suggestions for an autocomplete.
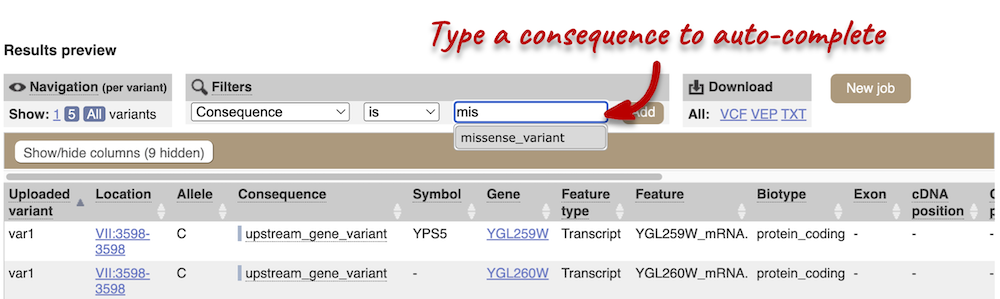
You can export your VEP results in various formats, including VCF. When you export as VCF, you’ll get all the VEP annotation listed under CSQ in the INFO column of the VCF file. After filtering your data, you’ll see that you have the option to export only the filtered data.
VEP in Ensembl Bacteria
In Ensembl Bacteria, launch the VEP tool and select the Bacteroides fragilis 638R genome. Use VEP to predict the effects of a C to A substitution at 3,835,830 bp. The variant in the Ensembl VEP default format is as follows:
Chromosome 3835830 3835830 C/A
Use the results to answer the following questions:
-
How many genes does the variant overlap? Do any of these genes have gene symbols?
-
What is the most common consequence of the variant?
-
What is the most severe consequence? What gene does it affect and what does it do?
Type Bacteroides fragilis 638R into the species search box and open its species page. Click on Variant Effect Predictor at the bottom right. In the input form, enter the following: Chromosome 3835830 3835830 C/A
Make sure you name your VEP job something descriptive, so it’s easier for you to find later on. Click Run to get the results.
- Find the number of overlapped genes in the Summary statistics. You can find their gene symbols under the Symbol column in the results table below.
The variant overlaps 10 different genes. 3 have the following symbols assigned to them: sgcG, groS and groL. 7 genes do not have a gene symbol.
- Sort the table by Consequence by clicking on the column name.
The most common consequence is downstream_gene_variant (n=8).
- You can find a list of calculated consequences sorted by severity in the Ensembl Variation documentation.
According to the calculated variant consequence list, the most severe consequence is missense_variant (moderate impact) on the groS gene (Ensembl gene ID ENSB:bOsvWlXx5gu4UXi). The gene codes for a 10 kDa chaperonin.
Comparative genomics
Demo: gene trees and homology predictions
Fungal Compara
Gene trees
Let’s look at the homologues of Saccharomyces cerevisiae R64-1-1 TAZ1 (gene stable ID: YPR140W). This gene is involved in stress response and conserved across different taxonomic domains. Search for the gene and go to the Gene tab.
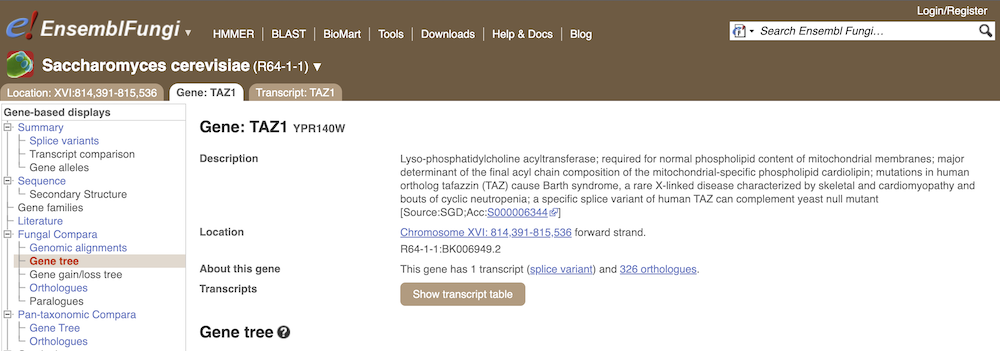
Click on Fungal Compara: Gene tree. This will display the current gene in the context of a phylogenetic tree used to determine orthologues and paralogues.
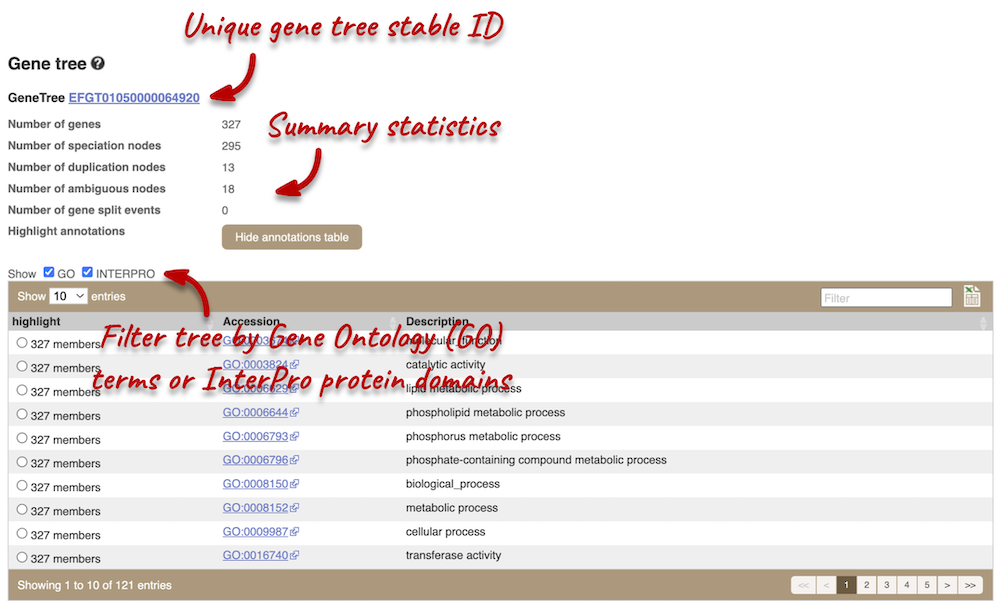
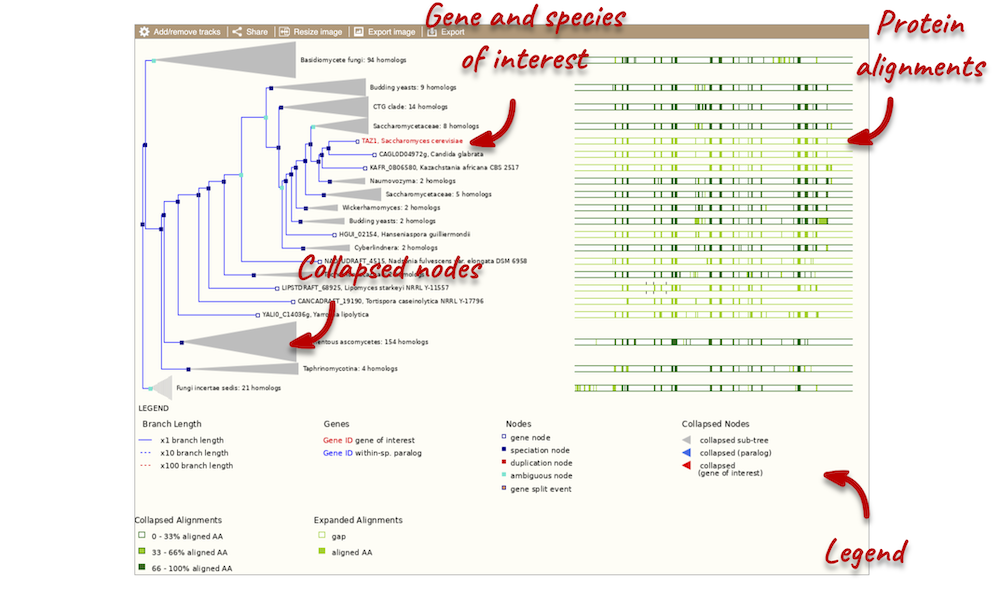
Funnels indicate collapsed nodes. Click on a node (coloured triangle) to open a menu. We can then see what type of node this is, some statistics and options to expand or export the sub-tree.

There are some quick filtering options below the image, where you can add paralogues, and quickly expand or collapse nodes.
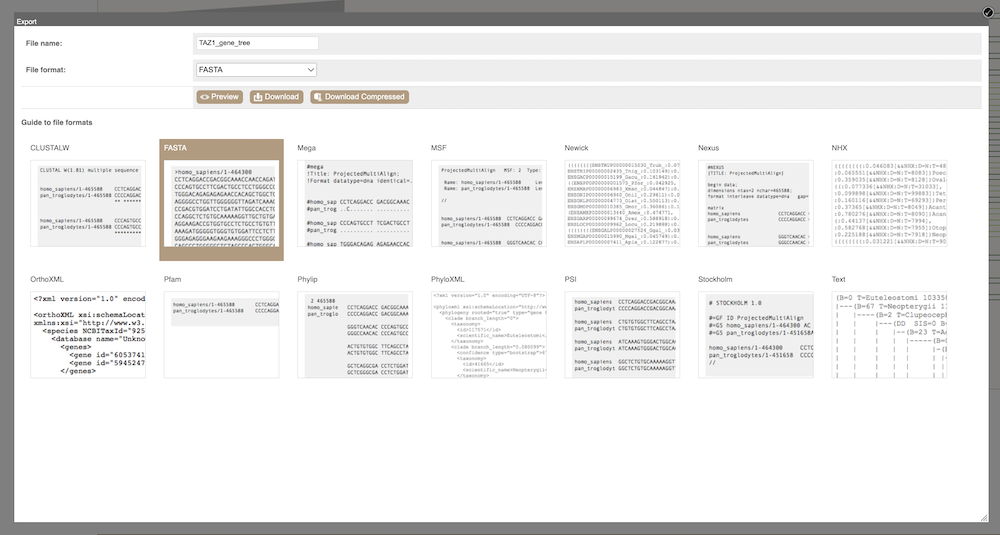
You can download the tree in a variety of formats. From the pop-up above you can click to export the sub-tree (everything to the right of the node). Alternatively, click on the Export icon ![]() in the bar at the top of the image to get a pop-up where you can choose your format. You can preview this file before you download.
in the bar at the top of the image to get a pop-up where you can choose your format. You can preview this file before you download.
Homologues
We can look at homologues in the Orthologues and Paralogues pages, which can be accessed from the left-hand menu. If there are no orthologues or paralogues, then the name will be greyed out. Click on Fungal Compara: Orthologues to see the orthologues available. In the first table, you will find a summary of orthologues by taxonomic group:

In the second table, you will find orthologue details per species:
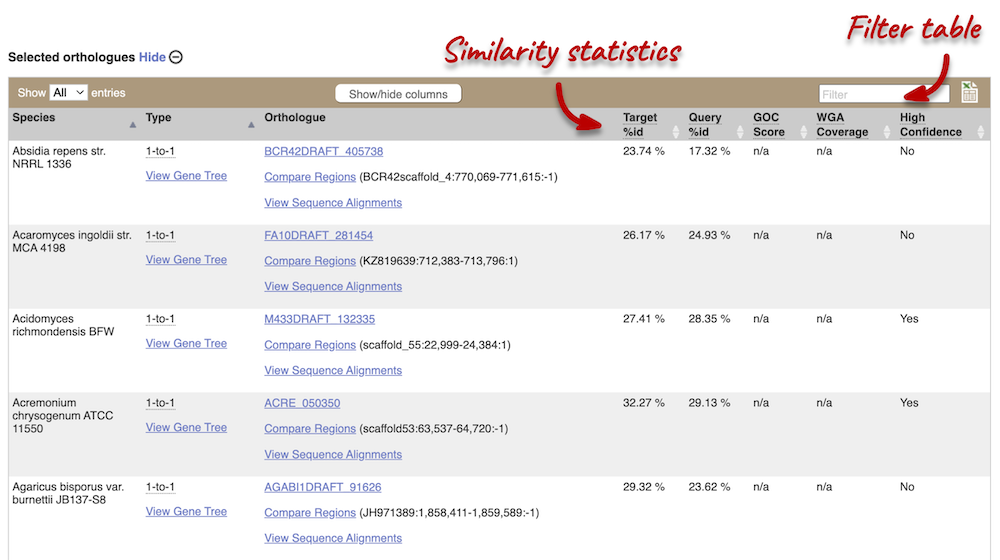
Scroll to the bottom of the page to see a list of the species that do not have any orthologues with TAZ1 in S. cerevisiae… there are a lot!

Pan-taxonomic Compara
S. cerevisiae is part of Pan-compara, which compares a subset of fungal species with species from other taxa, such as plants, bacteria and vertebrates. Click on Pan-taxonomic Compara: Orthologues. Let’s see if there are any orthologues of TAZ1 in plants. Click the Show details box for Plants.


Demo: Whole-genome alignments
Alignments in the Region in Detail view
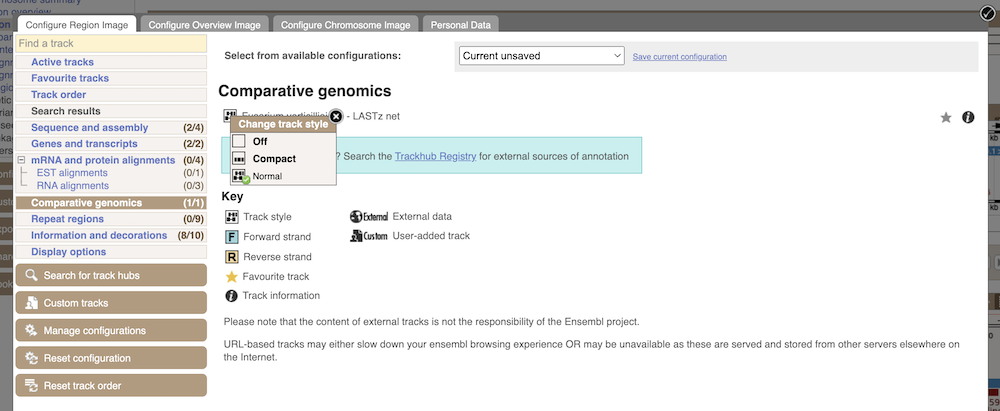
Let’s look at some of the comparative genomics views in the Location tab. Go to the region 2:438432-458358 in Fusarium solani. This region includes a number of genes and we want to find out if any regions align with Fusarium verticillioides. To do this, we need to look at a pairwise alignment between the two Fusarium species. We can look at individual species comparative genomics tracks in this view by clicking on Configure this page. In the Comparative genomics section, turn on the F. verticillioides track in the normal format.
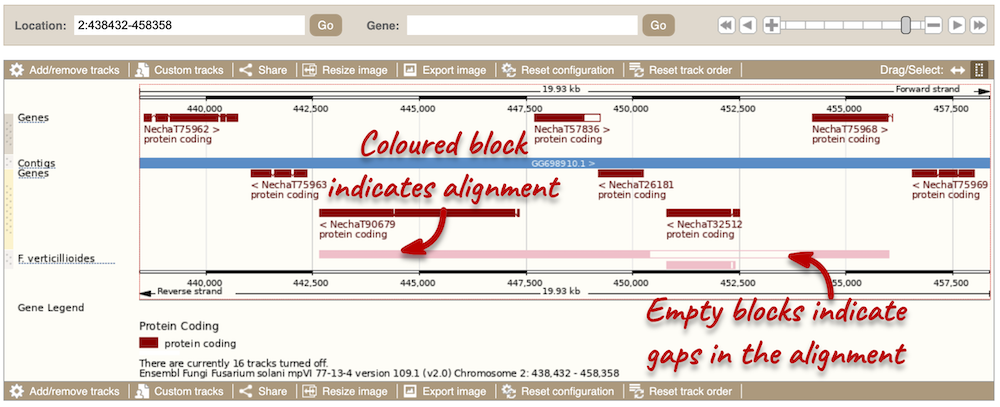
We can now see some pink alignments shown on the display. Alignments to the same chromosome are presented in a single row, and gaps in the alignment are shown by empty blocks. If there are alignments to multiple chromosomes in the aligned species, they are represented on different rows. From the track, we can see that the regions encoding genes NechaG90679, NechaG57836, NechaG26181, NechaG32512 and NechaG75968 align perfectly between the two genomes.
Sequence alignments
We can also look at the sequence alignment between the two species as text. Click on Comparative Genomics: Alignments (text) in the left-hand menu.
Click on Select an alignment to select a species you want to align. Let’s select F. solani and click Go. There are two blocks aligned of different lengths, some of which correspond to the region we just saw in the Region in detail view.
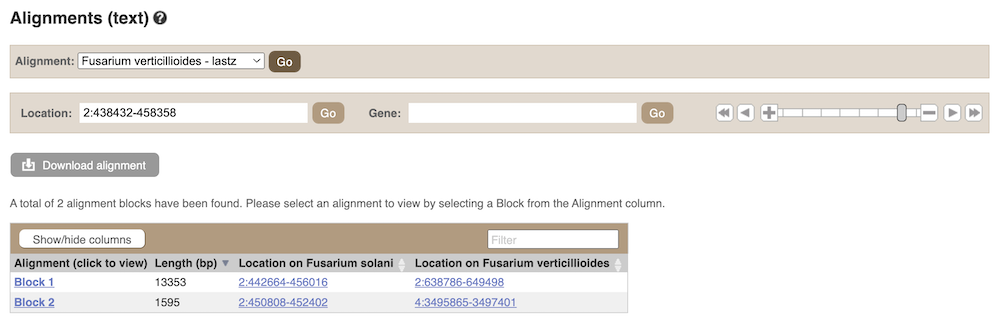
Click on Block 1.
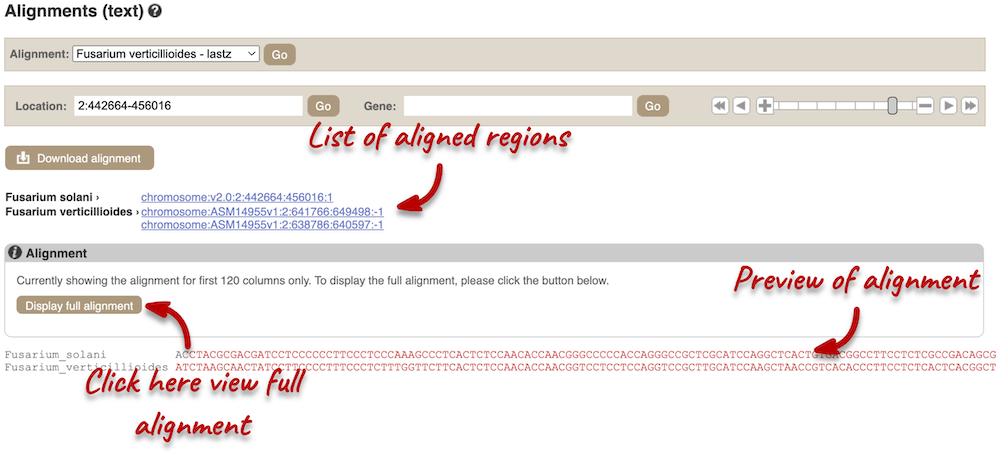
You will see a list of aligned regions, followed by the sequence alignment. Click on Display full alignment. Exons are shown in red. Click on Configure this page on the left. In the pop-up menu, you can turn on the options to view Show conservation regions and Mark alignment start/end. This will add highlights where the sequence matches.
Region comparison
To compare the two genomic regions visually, go to Region Comparison in the left-hand panel. To add species to this view, click on the Select species or regions button. Select F. verticillioides again then close the menu. This page, similar to the Region in detail view, shows the chromosome positions first. We can see the location of this alignment on chromosome 2 in F. solani. You can scroll down to the most detailed image to view aligned regions, which are highlighted and linked in green.
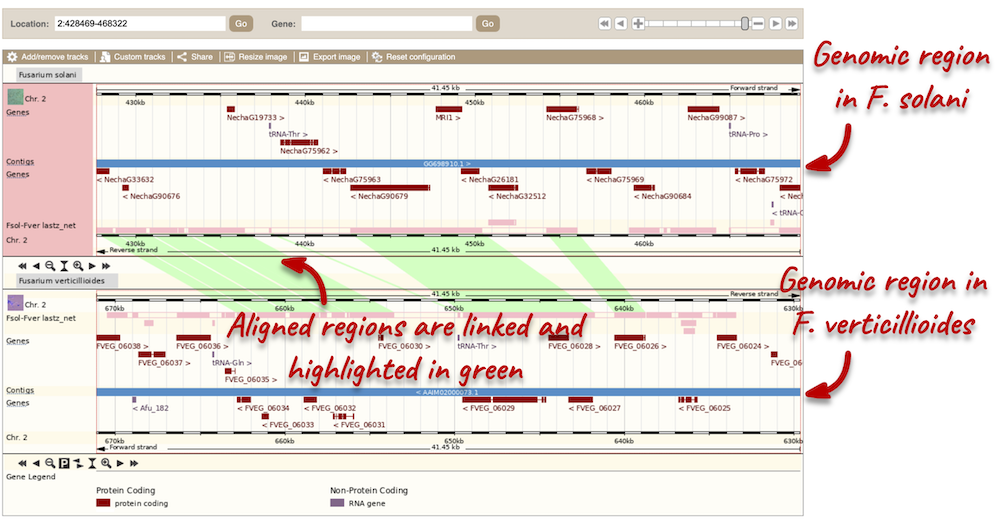
You can add data to both of these views with the same options you had in the Region in detail page. Click on Configure this page to open the menu.
Synteny
We can view large-scale syntenic regions from our chromosome of interest. Click on Synteny in the left-hand panel. Black linking lines indicate sequences are oriented in the same directed, red linking lines indicate the sequences are inverted.
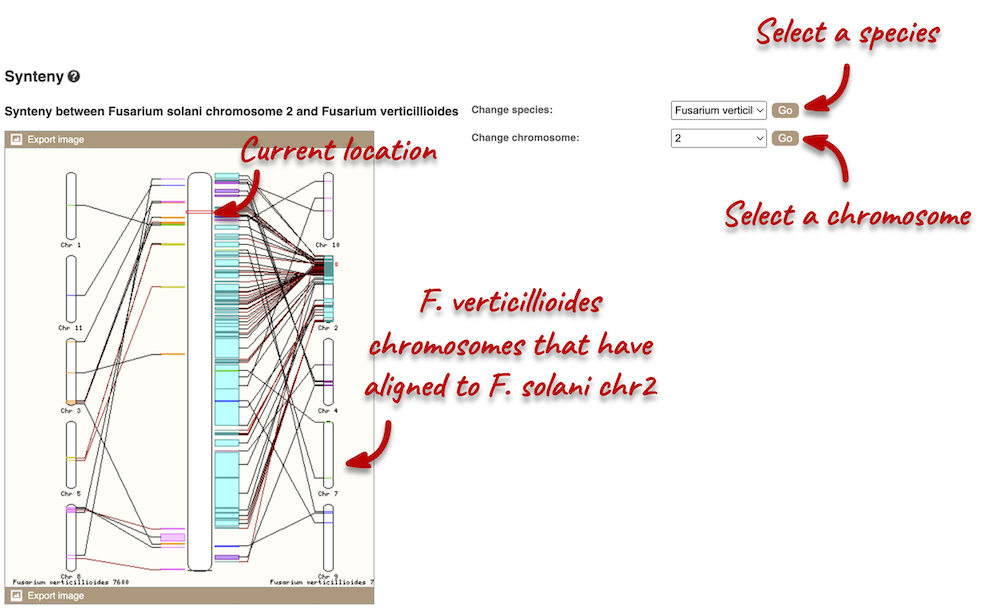
Pan-taxonomic comparative genomics data in Ensembl Bacteria
Bacillus subtilis subsp. subtilis str. 168 (GCA_000009045) is a model organism and often used in academic research and in the biotechnology industry as it can produce large amounts of important enzymes, like protease and amylase. It is part of Pan-taxonomic Compara in Ensembl Bacteria. We will use the sipT gene, a type I signal peptidase, as a reference to find the following information:
-
Find the Ensembl gene tree ID. How many speciation and duplication nodes does it have?
-
How many orthologues does B. subtilis str. 168 have? What type of orthologues are they?
-
Does it have an orthologue in Escherichia coli str. K-12 substr. MG1655? If so, what is the gene ID and coordinate in E. coli?
-
Export the protein alignment of the B. subtilis and E. coli orthologues. What are the different formats you can export the alignment as?
- Go to the Ensembl Bacteria homepage and enter
Bacillus subtilis subsp. subtilis str. 168 (GCA_000009045)in the Species search bar. In the species information page, entersipT. Click the gene ID BSU_14410. Under the Gene tab, click on Pan-taxonomic Compara: Gene Tree on the left.The Ensembl genetree ID is EGGT00050000013001. There are 92 speciation nodes and 35 duplication nodes.
- Go to Pan-taxonomic Compara: Orthologues on the left-hand panel. You can find the number and types of orthologues under the Summary of orthologues of this gene table.
The B. subtilis str. 168 sipT gene has 56 1-to-many and 15 many-to-many orthologues.
- Filter the Selected orthologues table by entering
_Escherichia coli_ str. K-12 substr. MG1655in the search bar in the top right-hand corner of the table.Yes, an orthologue is present in E. coli str. K-12 substr. MG1655. The gene ID is b2568 and the its coordinate is 2,704,335-2,705,309.
- Click on View Sequence Alignment in the Orthologue column. Select View Protein Alignment from the pop-up menu. Click on the Download homology button.
Depending on your downstream analyses you may choose to export the alignment in a particular format. In Ensembl, you can export the alignment in the following formats: ClustalW, FASTA, Mega, MSF, Nexus, OrthoXML, Pfam, Phylip, PhyloXML, PSI and Stockholm.
Custom data
Demo: Upload small files
We have some patients that present with microcephaly and developmental delay. They all have large scale deletions on chromosome five:
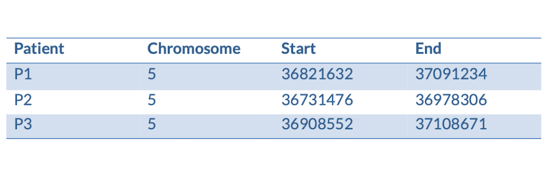
We can turn them into a BED file and view them in the genome browser:
chr5 36821632 37091234 P1
chr5 36731476 36978306 P2
chr5 36908552 37108671 P3
You can add data from a Region in Detail page by clicking on the Custom tracks button at the left. Alternatively, go to a species homepage and click on Display your data in Ensembl.

A menu will appear:
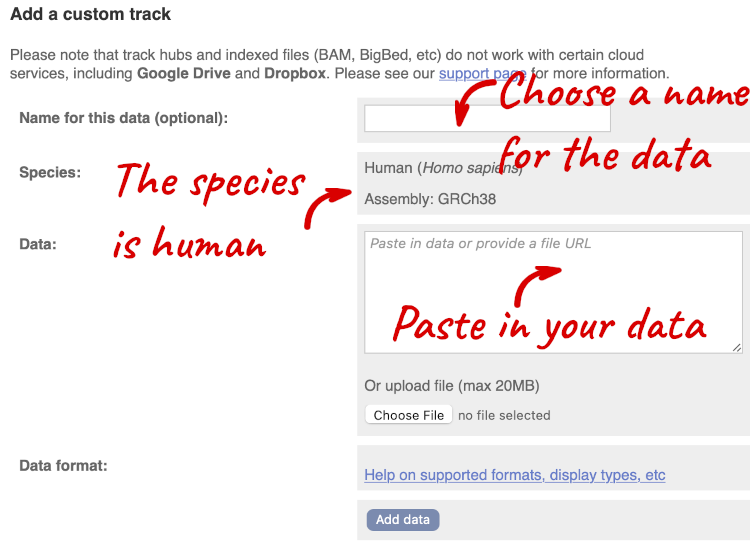
The interface detects file types if you upload or attach a file. When you paste in your data, it can’t do this so we have to tell it what our file type is. It will give you an option where you can select BED.
Click Add data.
You should get to a dialogue box telling you your upload has been successful.
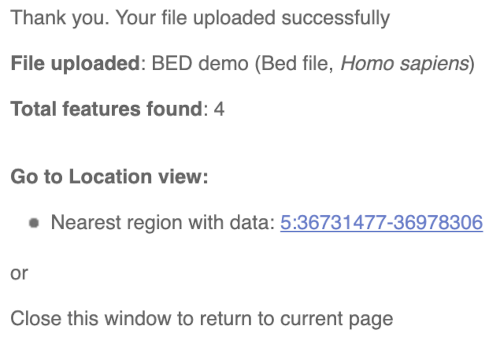
Click on the genomic coordinates link to go to the nearest region with data.
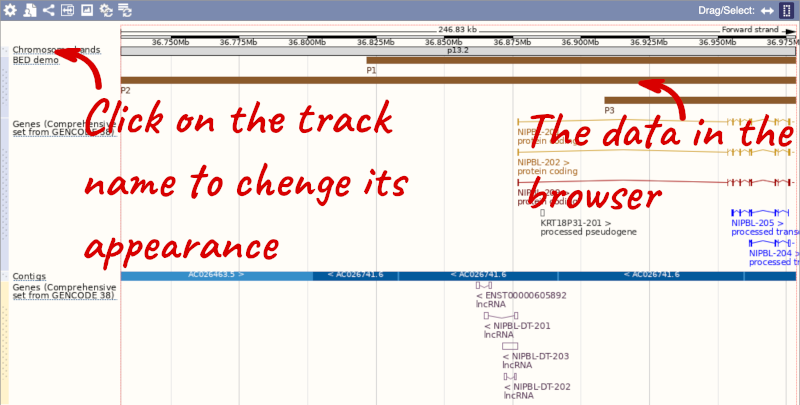
To have a look at the file, click on Custom tracks.
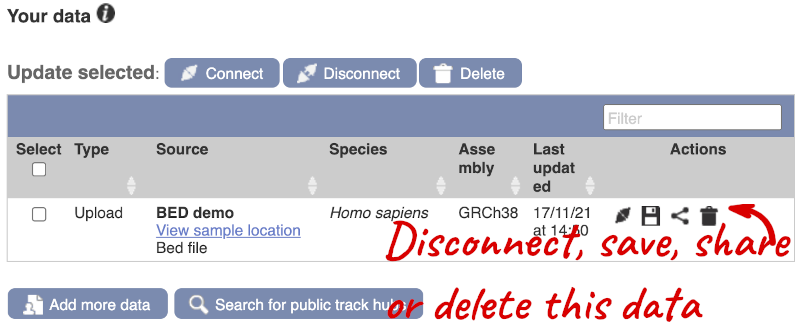
If you’ve got an Ensembl account, you can save this data to your account. Accounts are free to set up and allow you to save configurations and data, and share with groups.
Demo: Attach URLs of large files
Larger files, such as BAM files generated by NGS, need to be attached by URL. I’ve put a BAM file of human chromosome 20 RNASeq data online at: http://ftp.ebi.ac.uk/pub/databases/ensembl/training/emily_BAM/
Let’s take a look at the folder.
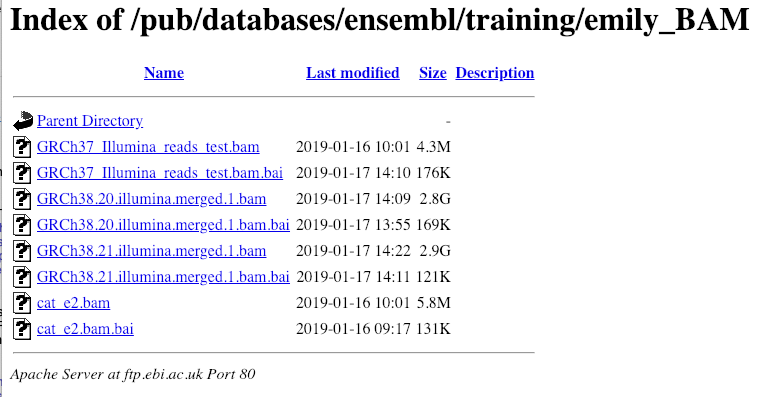
Here you can see a number of BAM files (.bam) with corresponding index files (.bam.bai). We’re interested in the files GRCh38.20.illumina.merged.1.bam and GRCh38.20.illumina.merged.1.bam.bai. These files are the BAM file and the index file respectively. When attaching a BAM file to Ensembl, there must be an index file in the same folder.
To attach the file, click on Custom tracks, then click on Add more data to add a new track.
We get to the same dialogue box as before. This time we’ll name our data Illumina reads.
Paste in the URL of the BAM file itself (http://ftp.ebi.ac.uk/pub/databases/ensembl/training/emily_BAM/GRCh38.20.illumina.merged.1.bam).
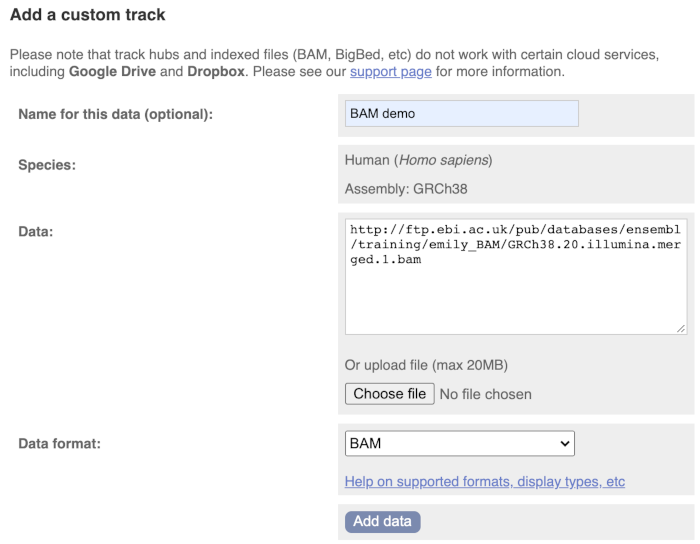
Since this is a file, the interface is able to detect the “.BAM” file extension, so automatically labels the format as BAM. Click on Add data and close the menu.
To see this data, jump to a region on chromosome 20. Let’s go to the region of the CDH22 gene. Search for the gene and click on the location.
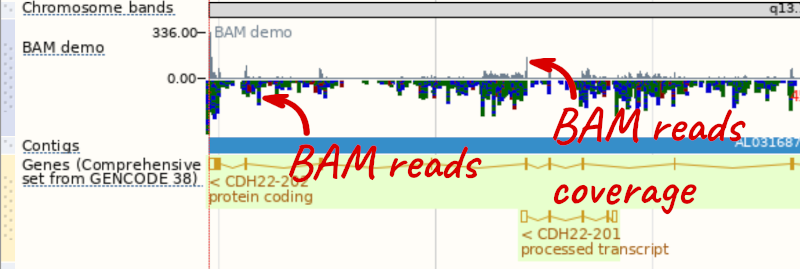
We can zoom in to see the sequence itself. Drag out boxes in the view to zoom in, until you see a view like this. Alternatively, jump to location 20:46241000-46241030.

Demo: Track hub registry
Our regulatory data incorporates data from sources such as ENCODE, Blueprint, and Roadmap Epigenomics. To see the data directly from these sources, you can add track hubs.
You can search for track hubs to add in different ways:
- Search for track hubs in the Track Hub Registry and choose to add them to your genome browser of choice.
- Search the track hub registry using the Track Hub Registry interface in Ensembl (there is a link from the homepage).
We will now add the track hub containing data from the Blueprint project.
You can add track hubs to view in Ensembl directly via the Track Hub Registry. Go to the Track Hub Registry homepage and search for blueprint.
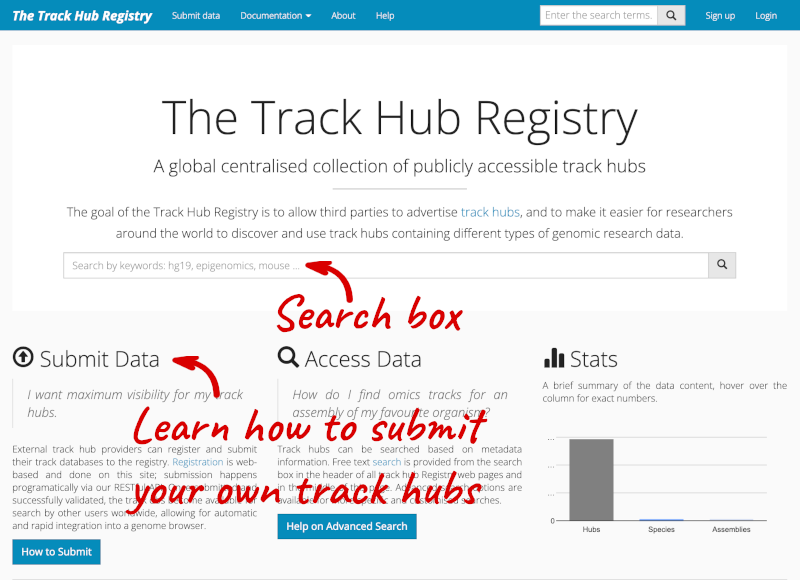
There are two results for the Blueprint Hub, one for adding the track hub to GRCh37 and one for adding it to GRCh38, plus one RNA-seq alignment hub.
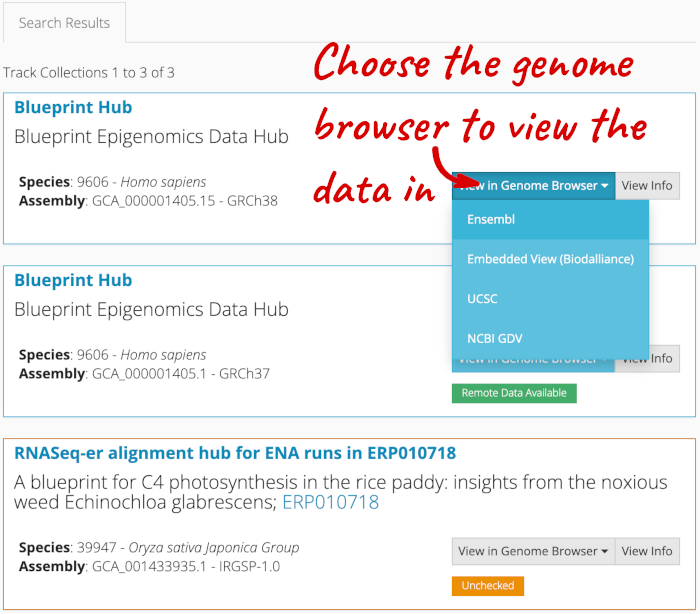
Alternatively, you can add track hubs by searching the Track Hub Registry through Ensembl. Click the Custom tracks -> Track Hub Registry Search in any region view within Ensembl.
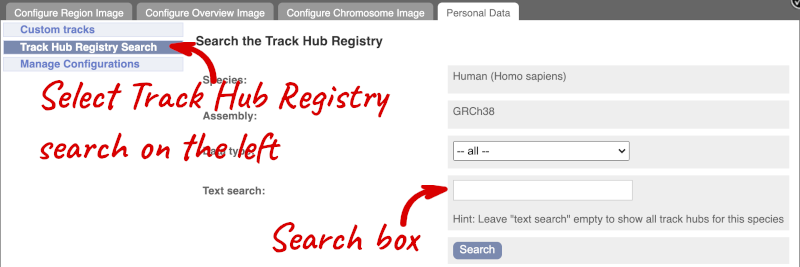
You can only find track hubs for the selected species and assembly denoted in the search box.
Search for blueprint.
Click Attach this hub in the search results page.
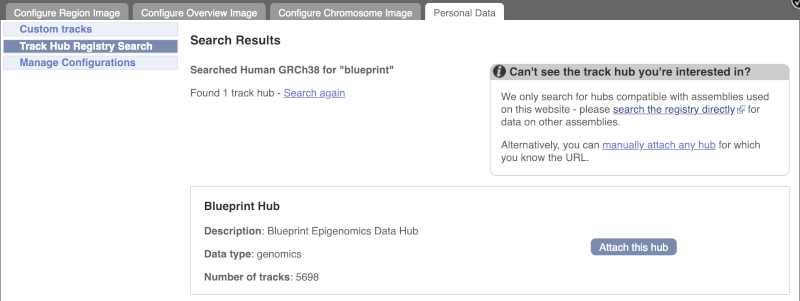
Track Hubs often contain vast amounts of data, which can slow Ensembl down, so only add them if you need them, and trash them when you are finished with them.
Go to Configure this Page to see that a new category has been added to your menu.
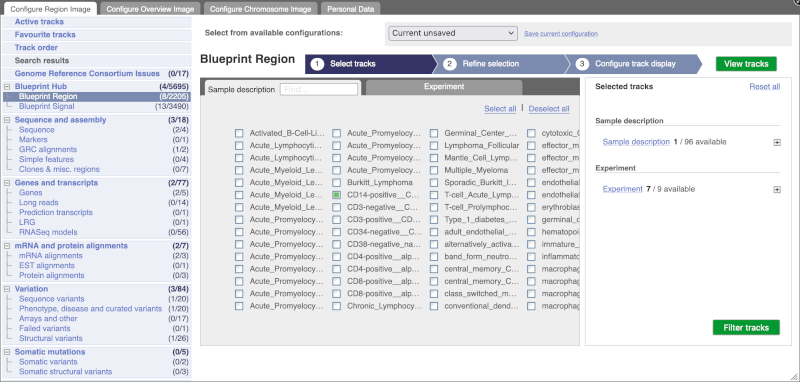
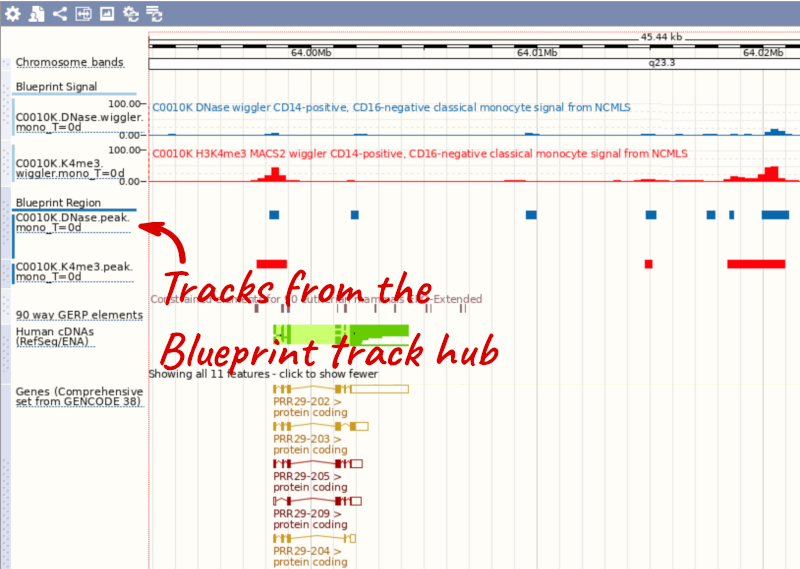
You can add tracks to the Region in Detail view using the matrix.
Adding Wiggle files to Ensembl Bacteria
Upload the GD_wiggle.wig file to the Gluconacetobacter diazotrophicus PA1 5 (GCA_000021325) genome in Ensembl Bacteria. View this track across the region Chromosome:2884000-2898000. What is the highest score in this region?
Go to Ensembl Bacteria and put Gluconacetobacter diazotrophicus PA1 5 into the Search for a genome box. Select Gluconacetobacter diazotrophicus PA1 5 (GCA_000021325) to go to the species homepage.
Select Display your data in Ensembl Bacteria to get to the custom track menu. Select Choose file and select the file location. The file type should be automatically selected. Click Add data.
Click on the Nearest region with data in the results page. From the region page you reach, put the coordinates Chromosome:2884000-2898000 into the Location box to jump to the region.
The highest score is 99 and it overlaps the ACI52364 transcript.







