Filter Events by Year
Ensembl Browser Workshop - Alliance Bioversity International - CIAT
Course Details
- Lead Trainer
- Aleena Mushtaq
- Event Date
- 2024-07-11
- Location
- Cali, Colombia
- Description
- Work with the Ensembl Outreach team to get to grips with the Ensembl browser, accessing gene, variation and comparative genomics data.
- Survey
- Ensembl Browser Workshop - Alliance Bioversity International - CIAT Feedback Survey
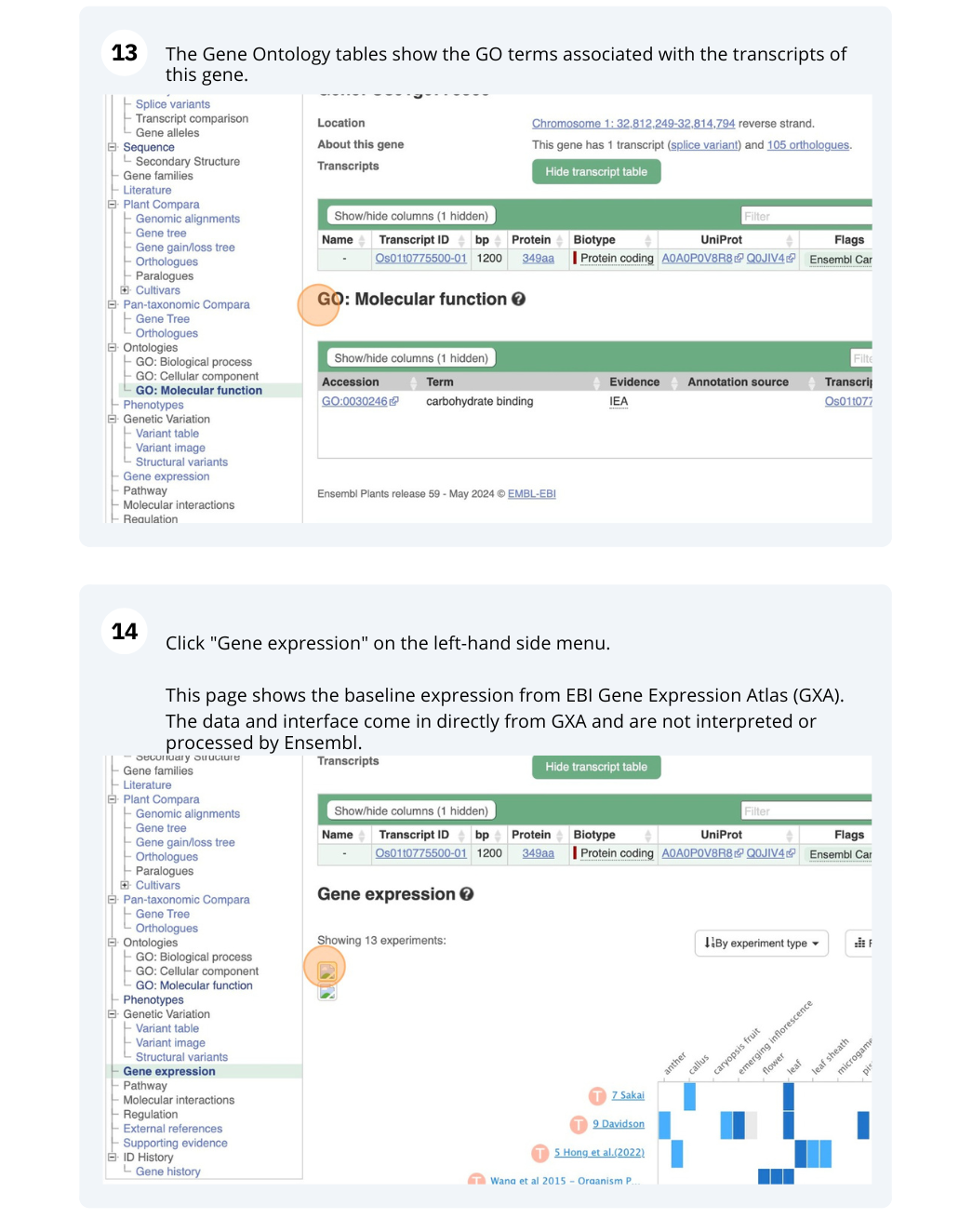
Demos and exercises
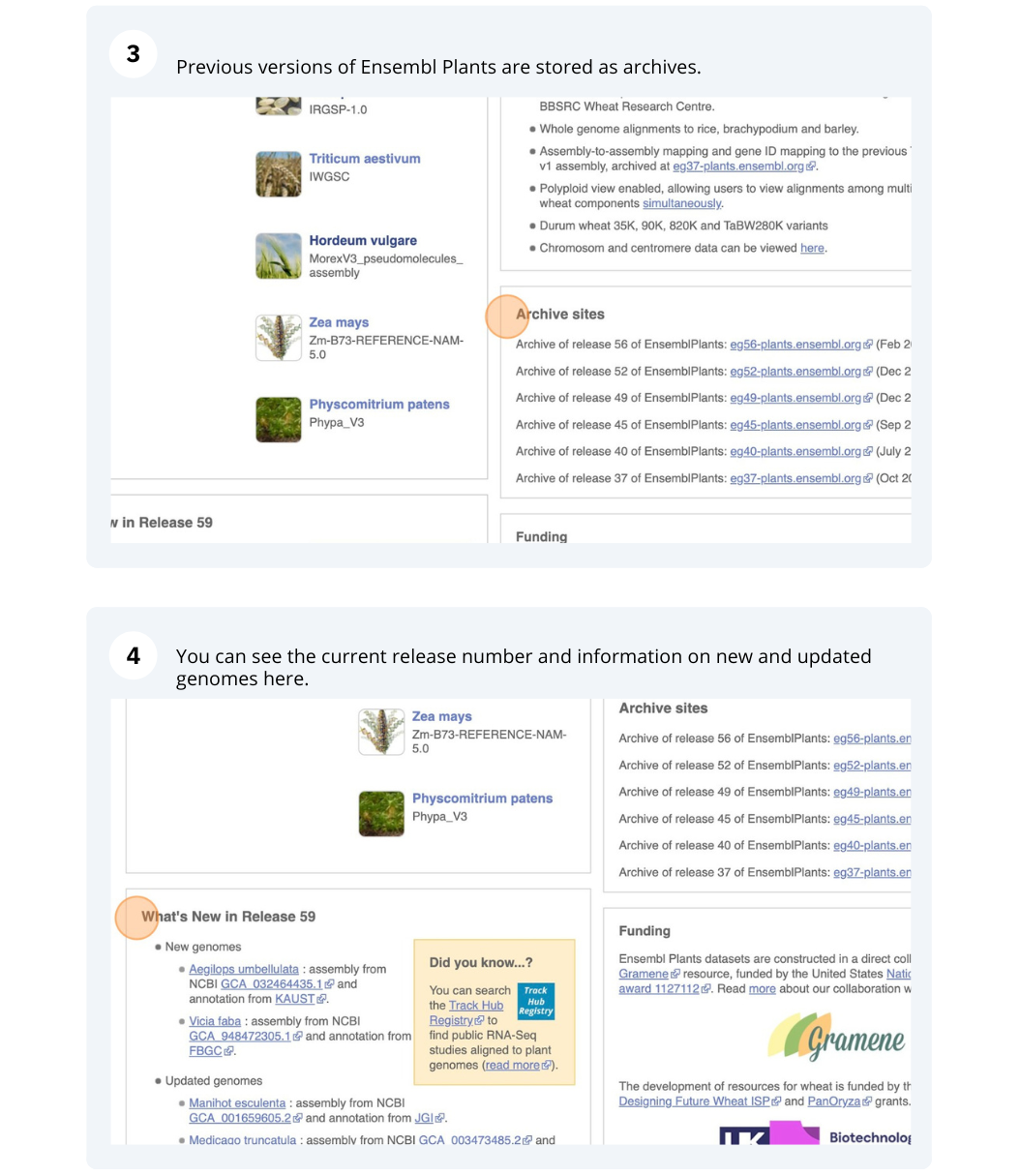
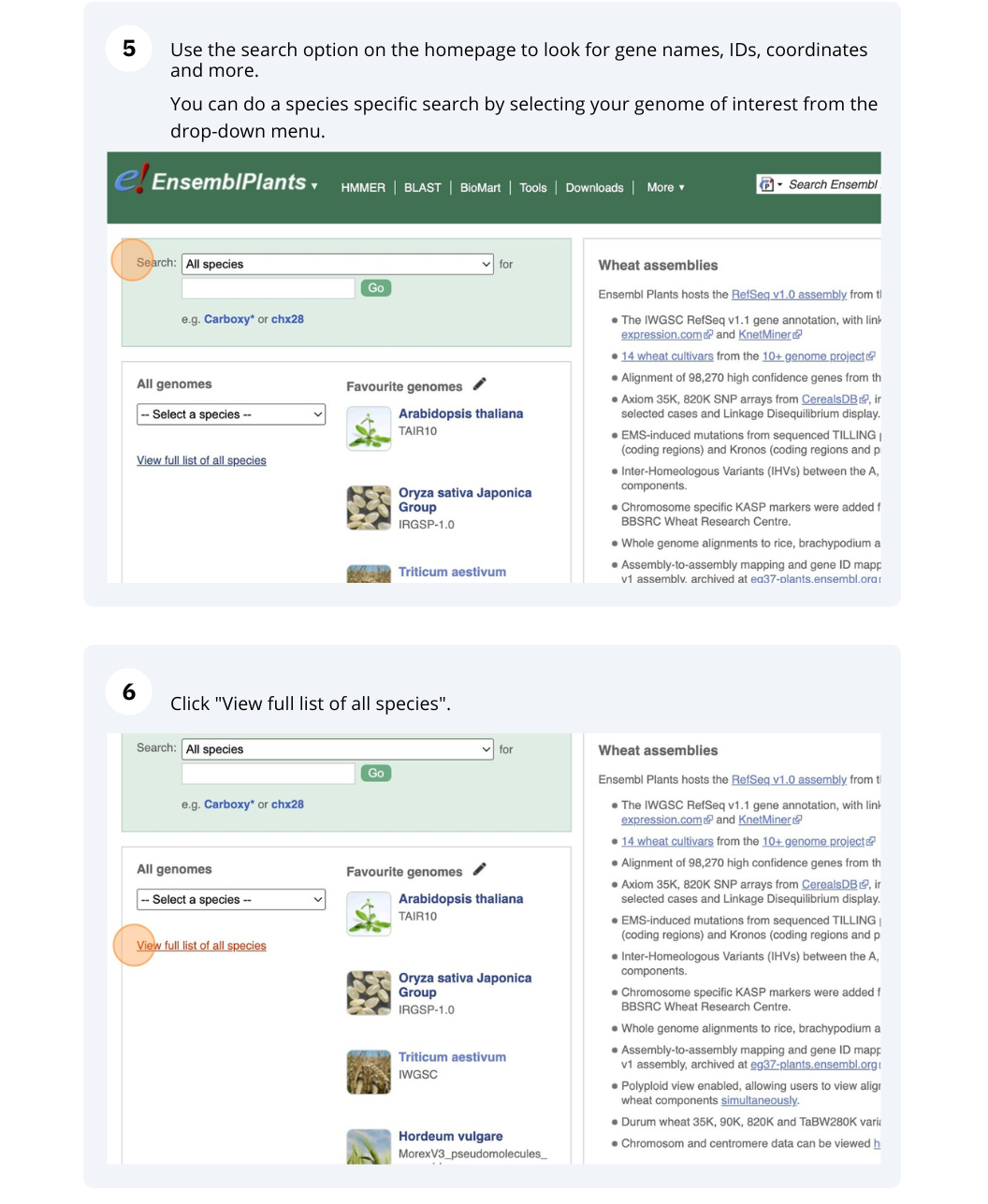
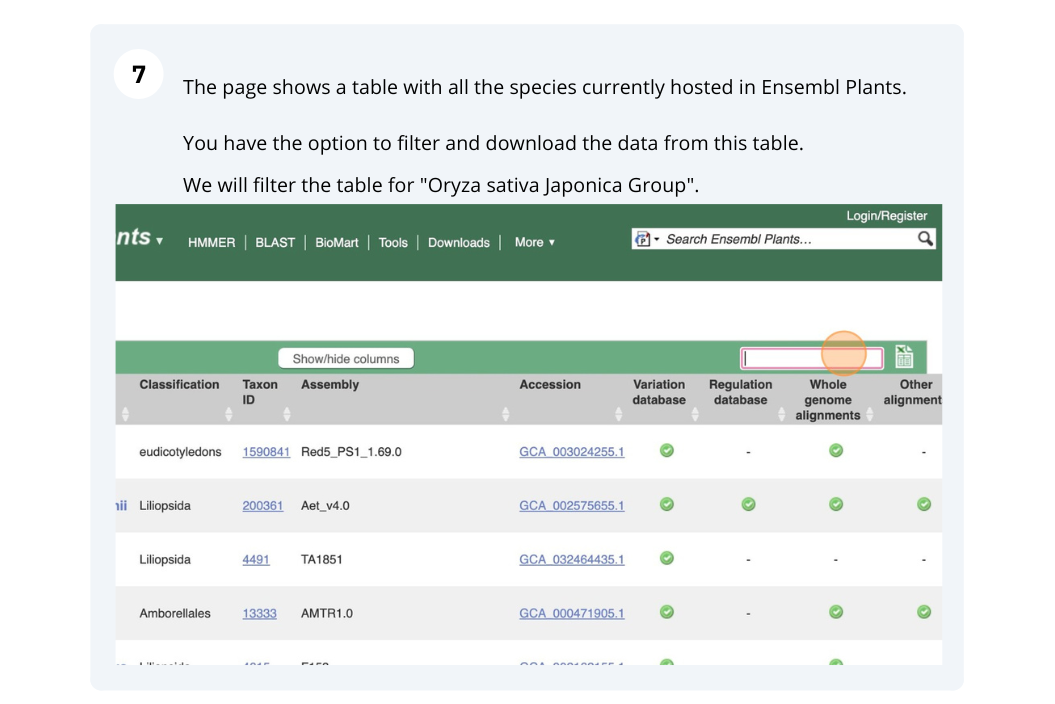
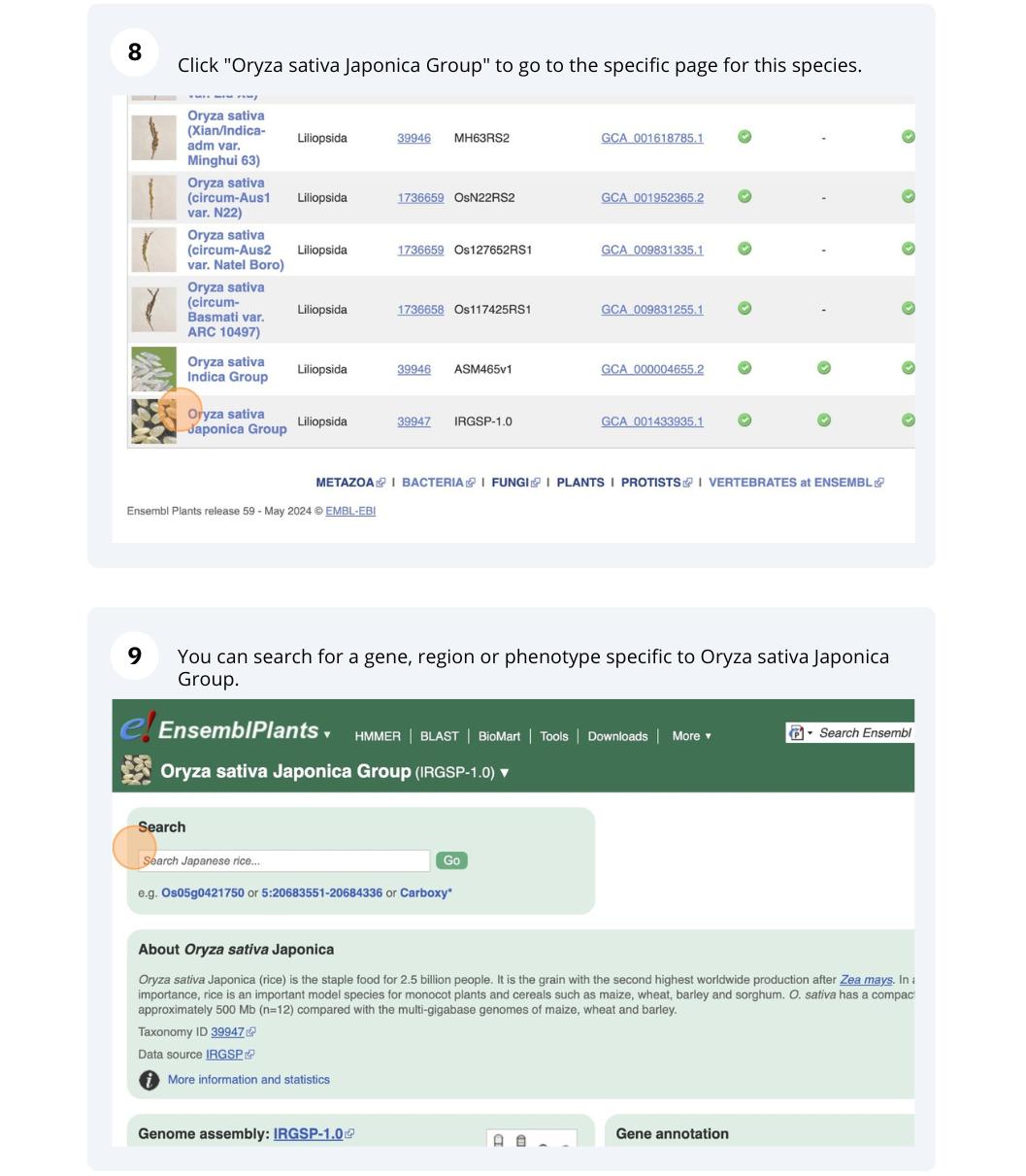
Ensembl Plants species
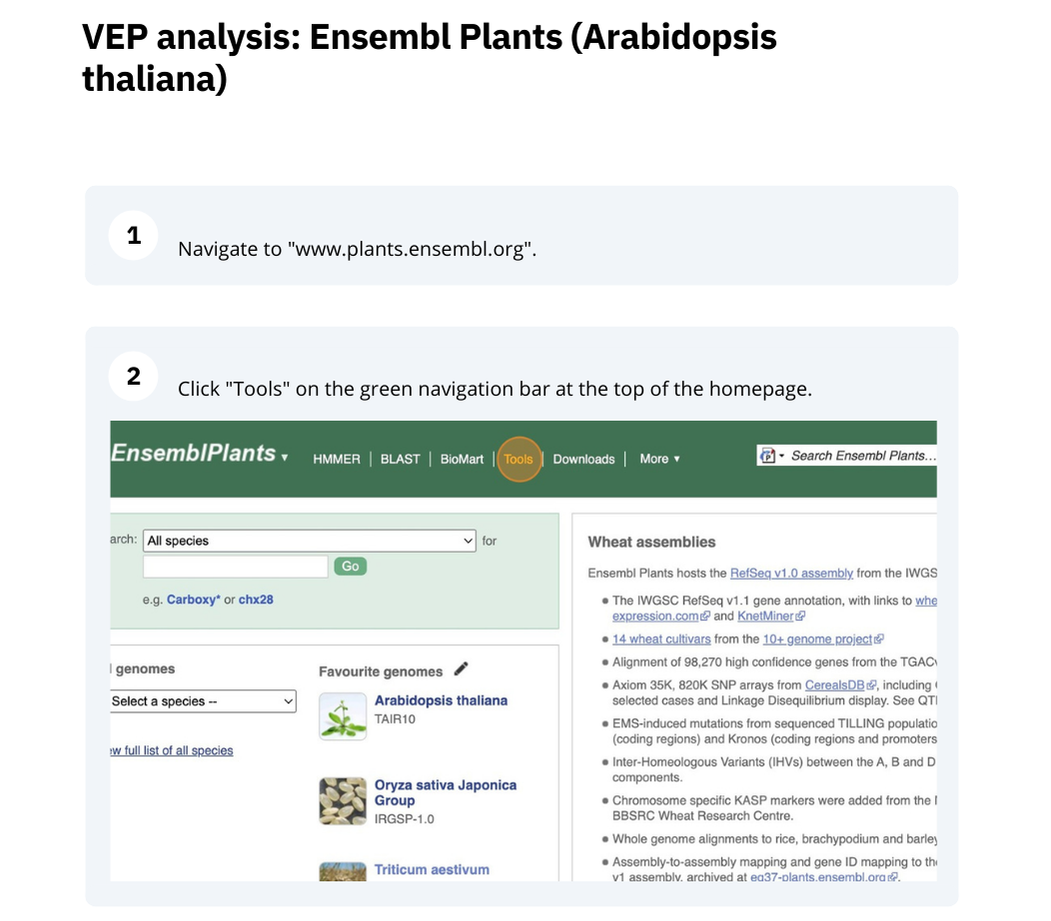
The front page of Ensembl Metazoa is found at www.plants.ensembl.org/. It contains lots of information and links to help you navigate Ensembl Plants.

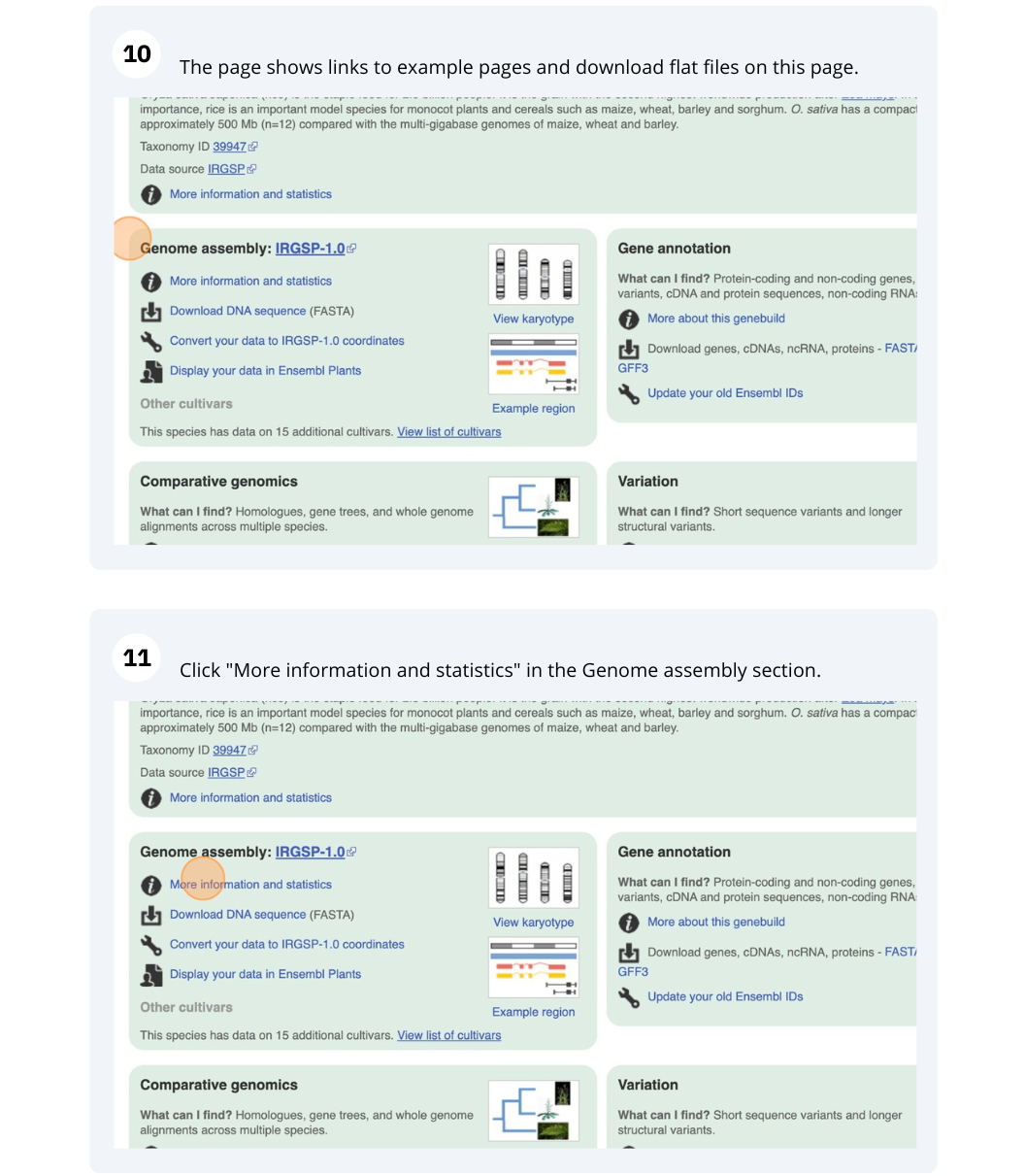
Finding information about Arabidopsis thaliana
(a) Go to the species homepage for the plant Arabidopsis thaliana. What is the name of the genome assembly for Arabidopsis thaliana?
(b) Click on More information and statistics. How long is the Arabidopsis thaliana genome (in bp)? How many genes and transcripts have been annotated?
(a) Go to plants.ensembl.org and select Arabidopsis thaliana from the species list.
The assembly is TAIR10.
(b) Click on More information and statistics. Statistics are shown in the tables on the left.
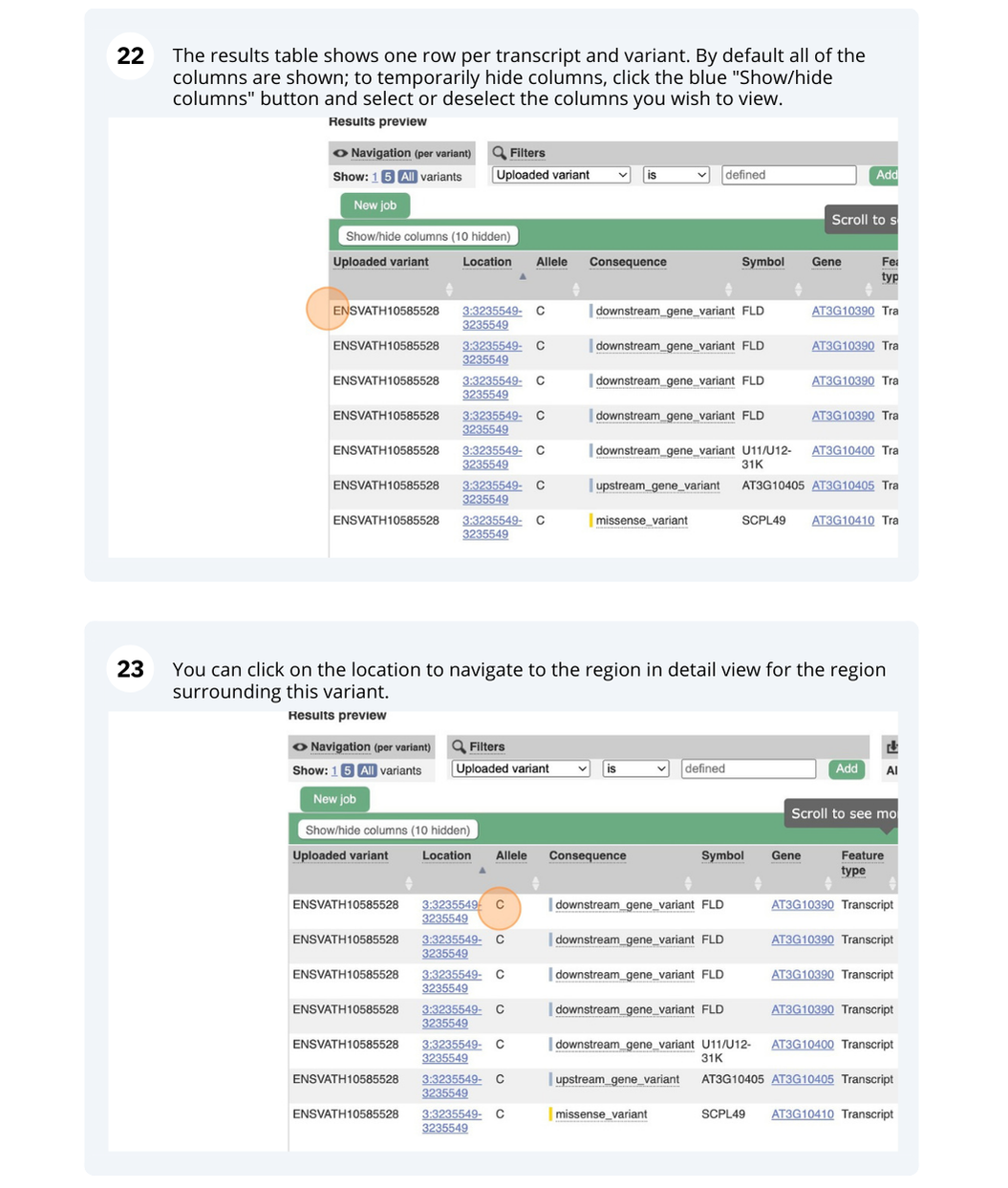
The length of the genome is 119,667,750 bp.
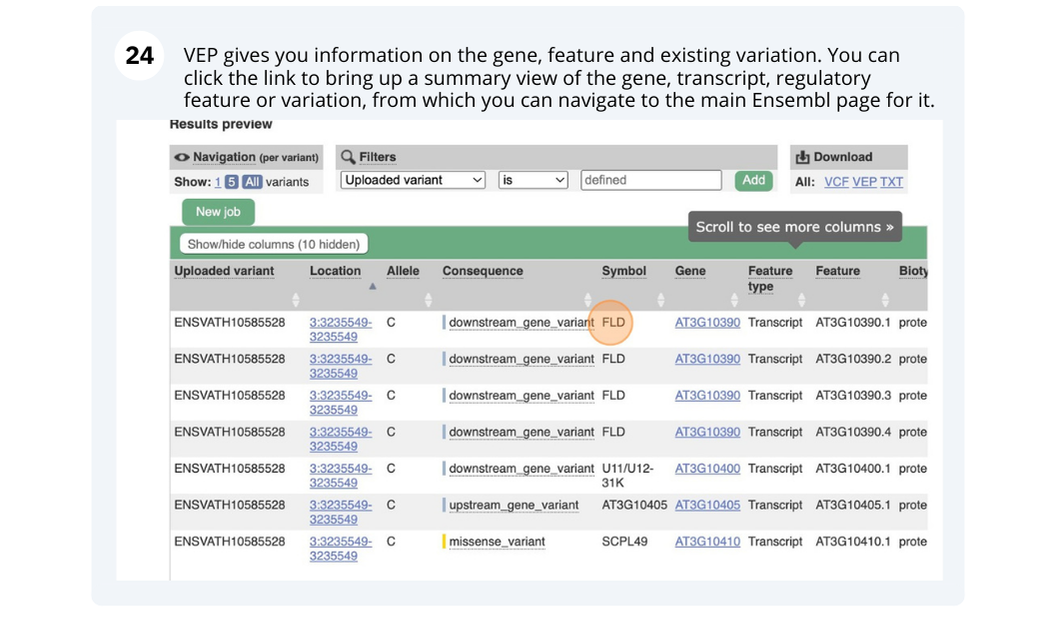
There are 27,655 coding genes and 54,013 transcripts.
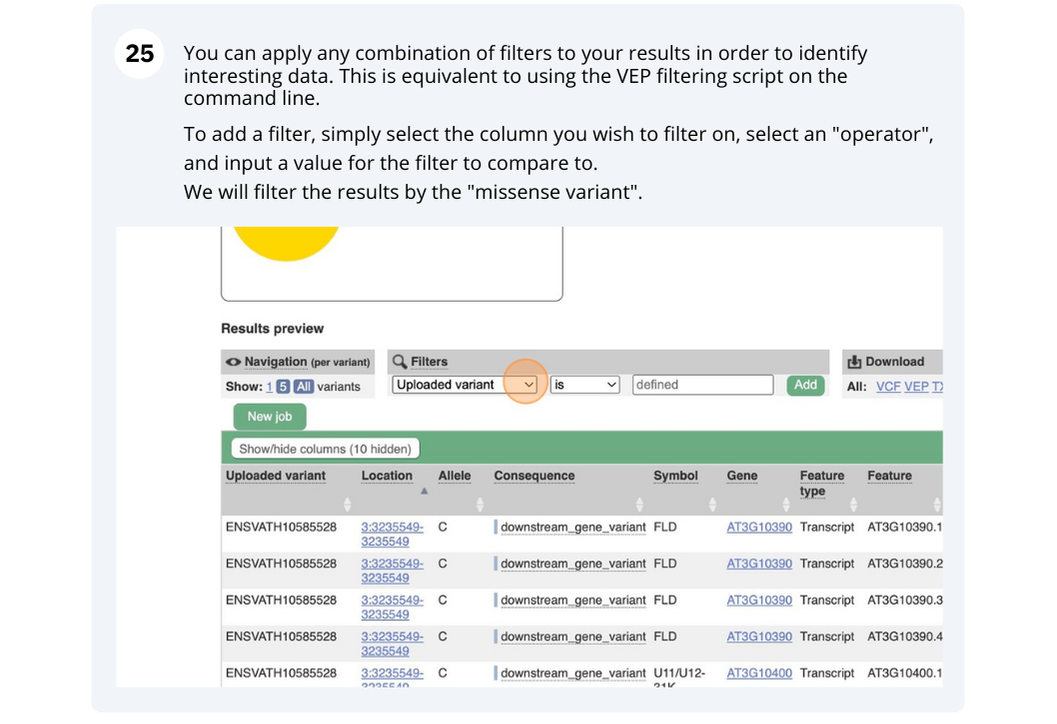
Mosquito species
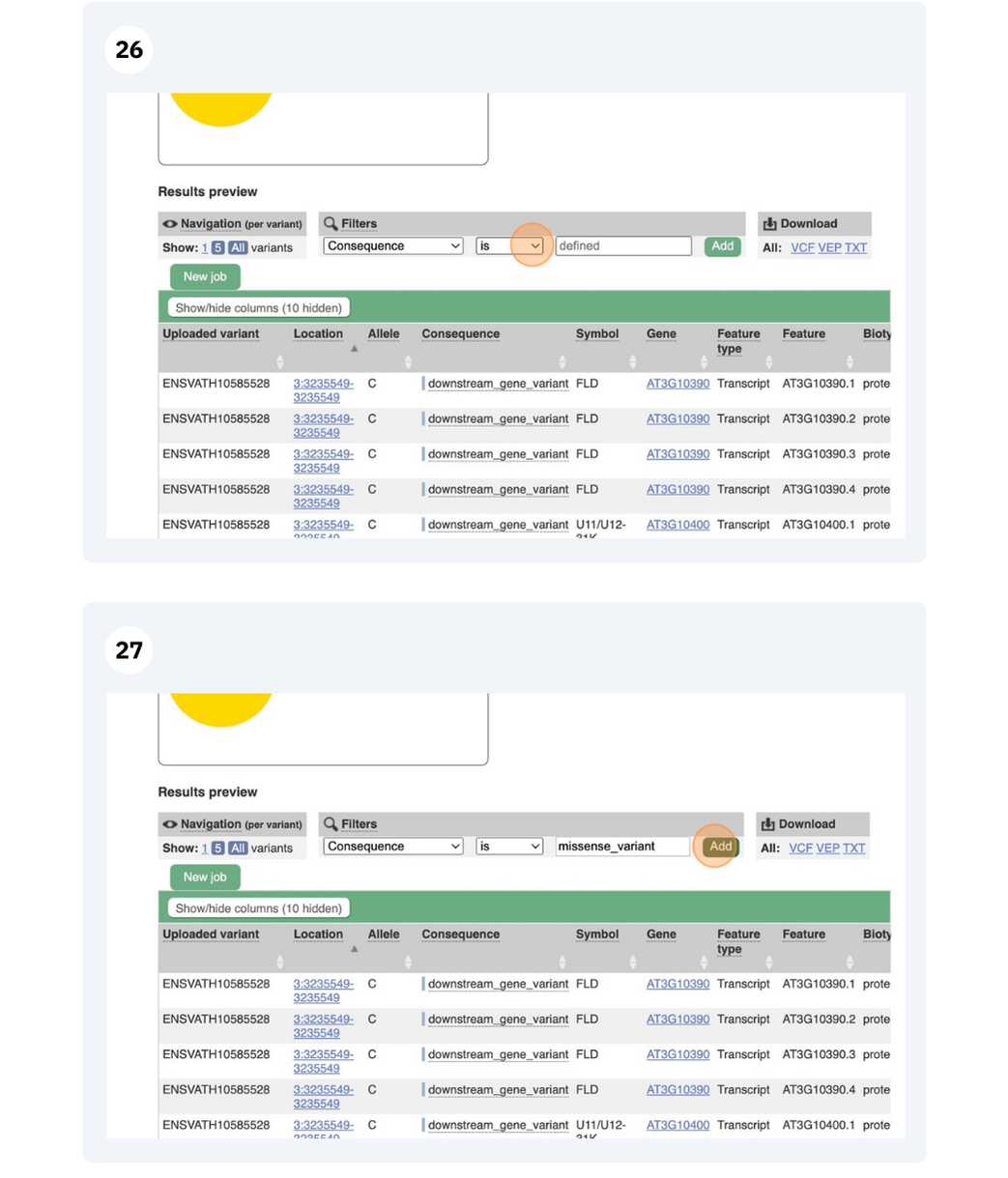
-
Go to Ensembl Metazoa. How many genomes relating to the genus Anopheles are there in Ensembl Metazoa?
-
When was the current Anopheles gambiae genome assembly last revised?
- Go to metazoa.ensembl.org. Open the drop-down list or click on View full list of all Ensembl Metazoa species. In a latin binomial species name, the first word represents the genus. Type Anopheles into the filter box in the top left to find all genomes with this word in the binomial.
There are 22 Anopheles genomes (some species are represented by more than one genome).
- Click on Anopheles gambiae (African malaria mosquito, PEST), and then on More information and statistics.
The assembly hosted is AgamP4 (INSDC Assembly GCA_000005575.1) which was revised in Feb 2006.
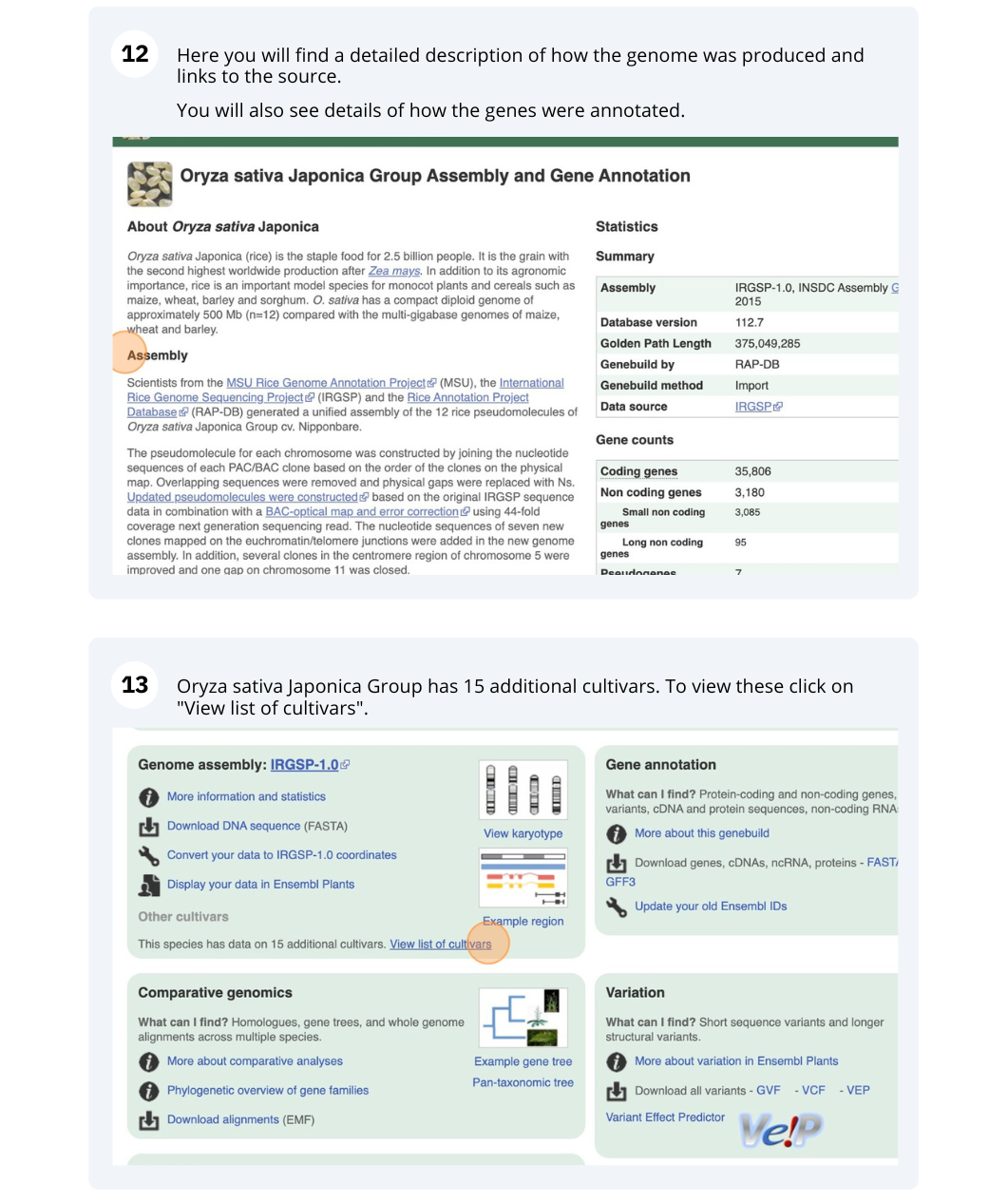
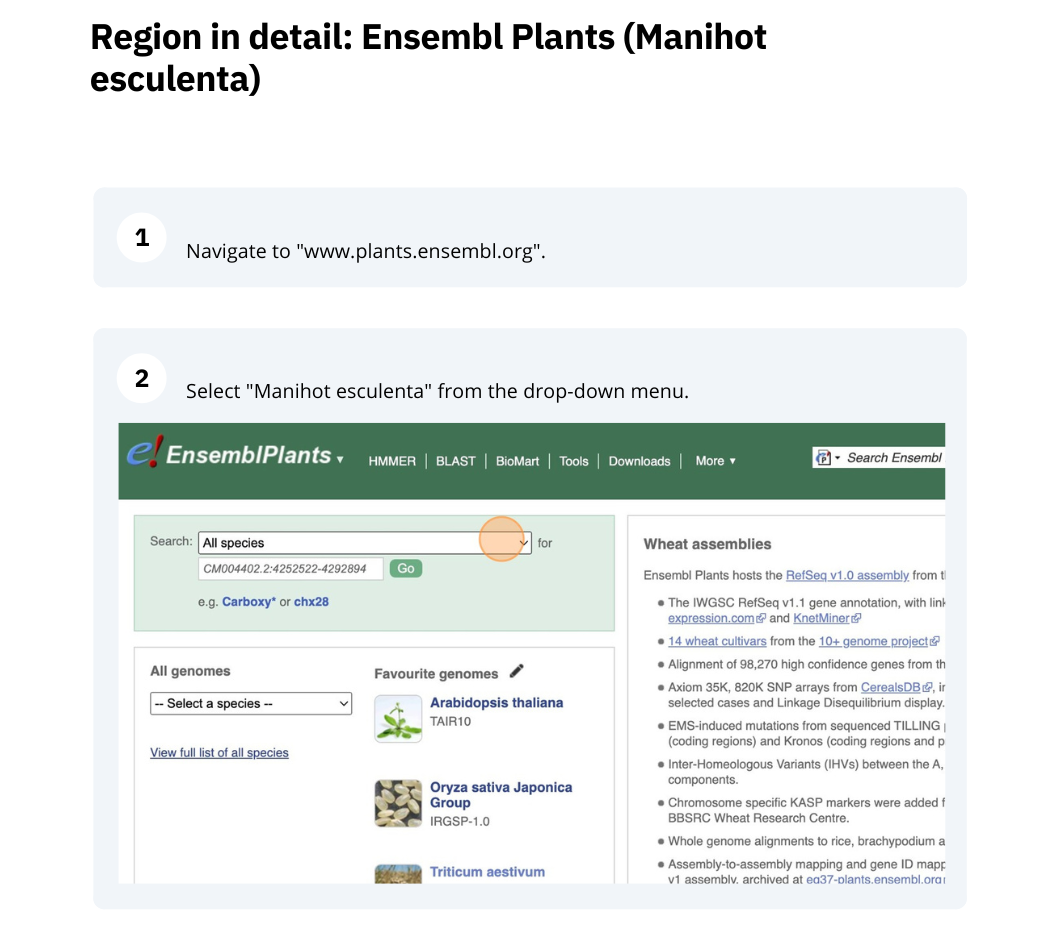
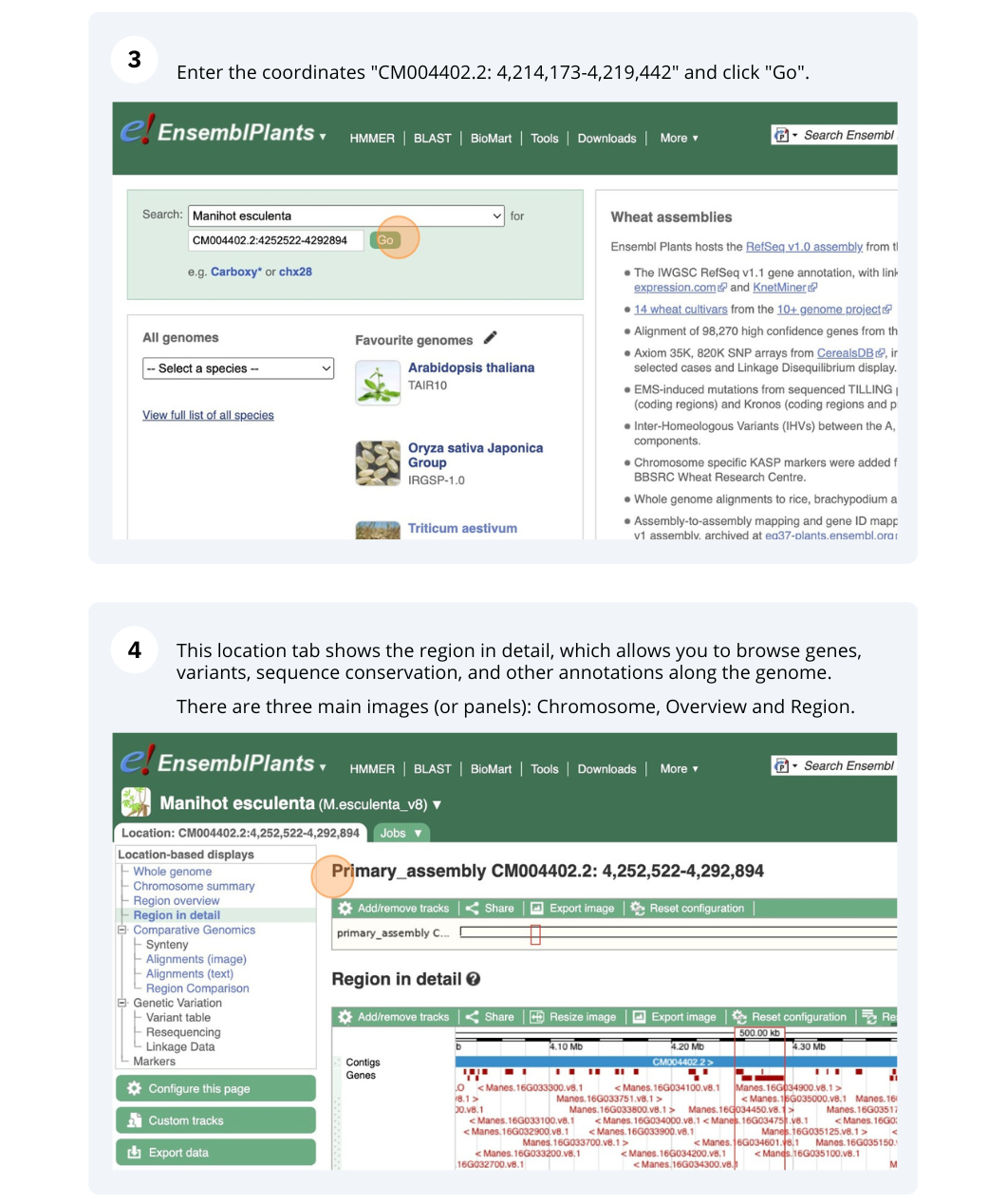
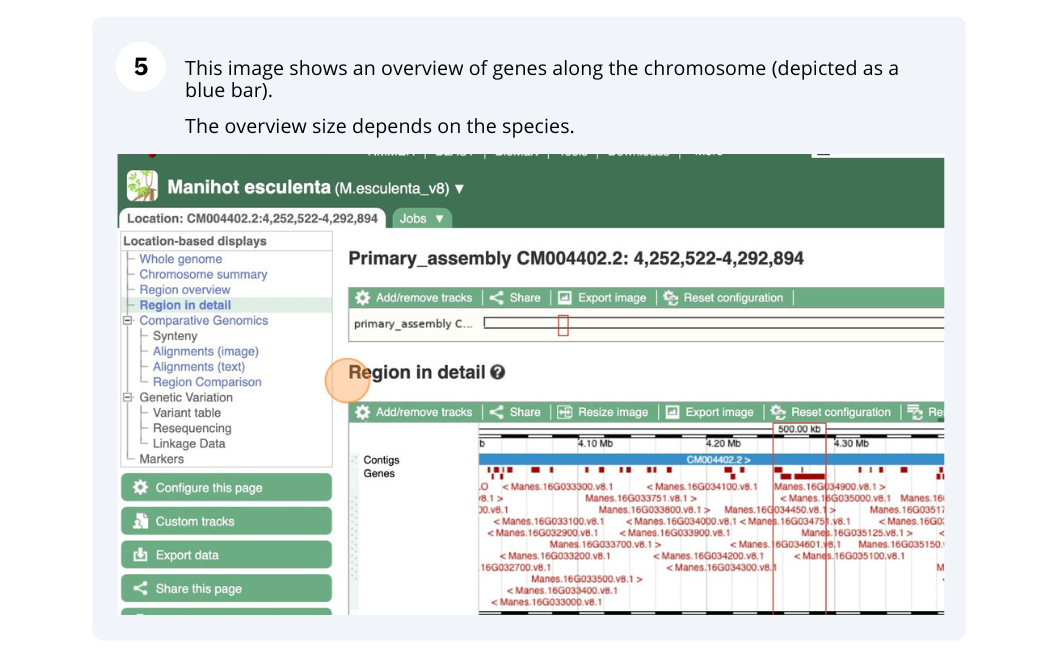
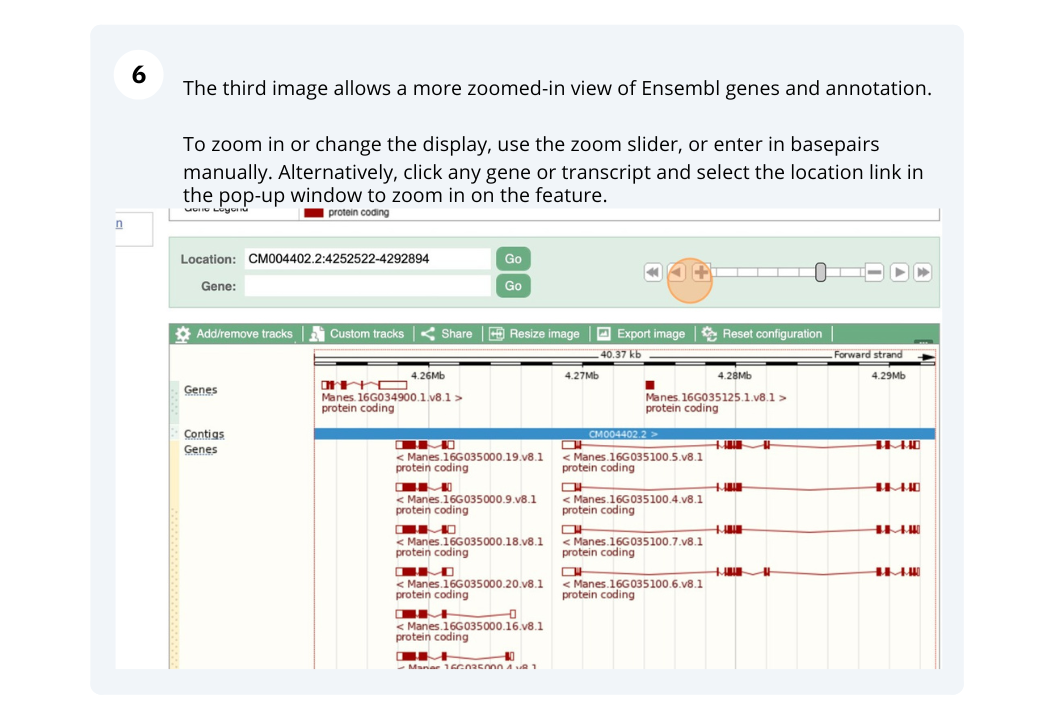
Ensembl Plants region in detail
We’re going to look at a region of the Manihot esculenta genome, CM004402.2: 4,214,173-4,219,442, and manipulate the view to see the data we are interested in.

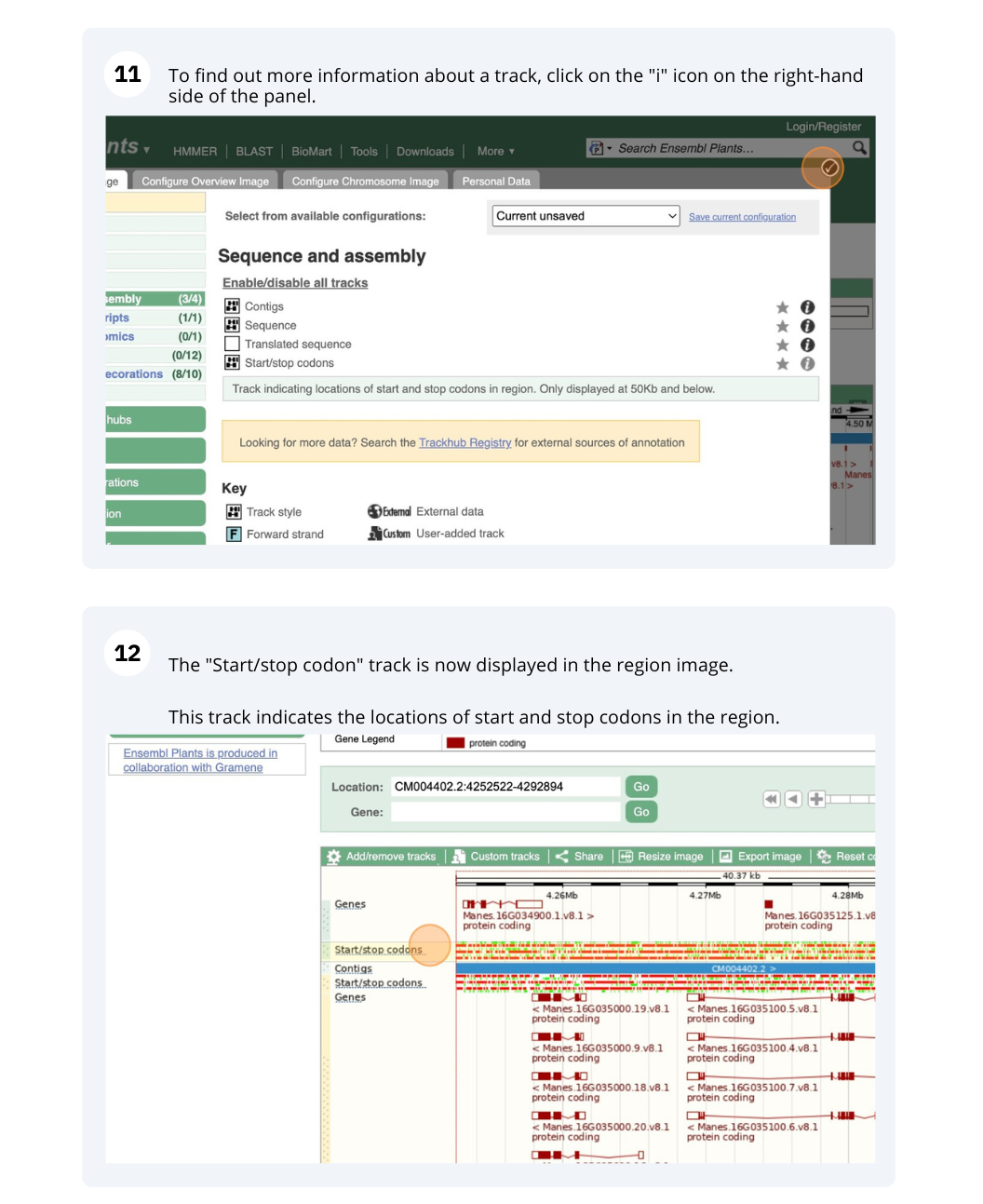
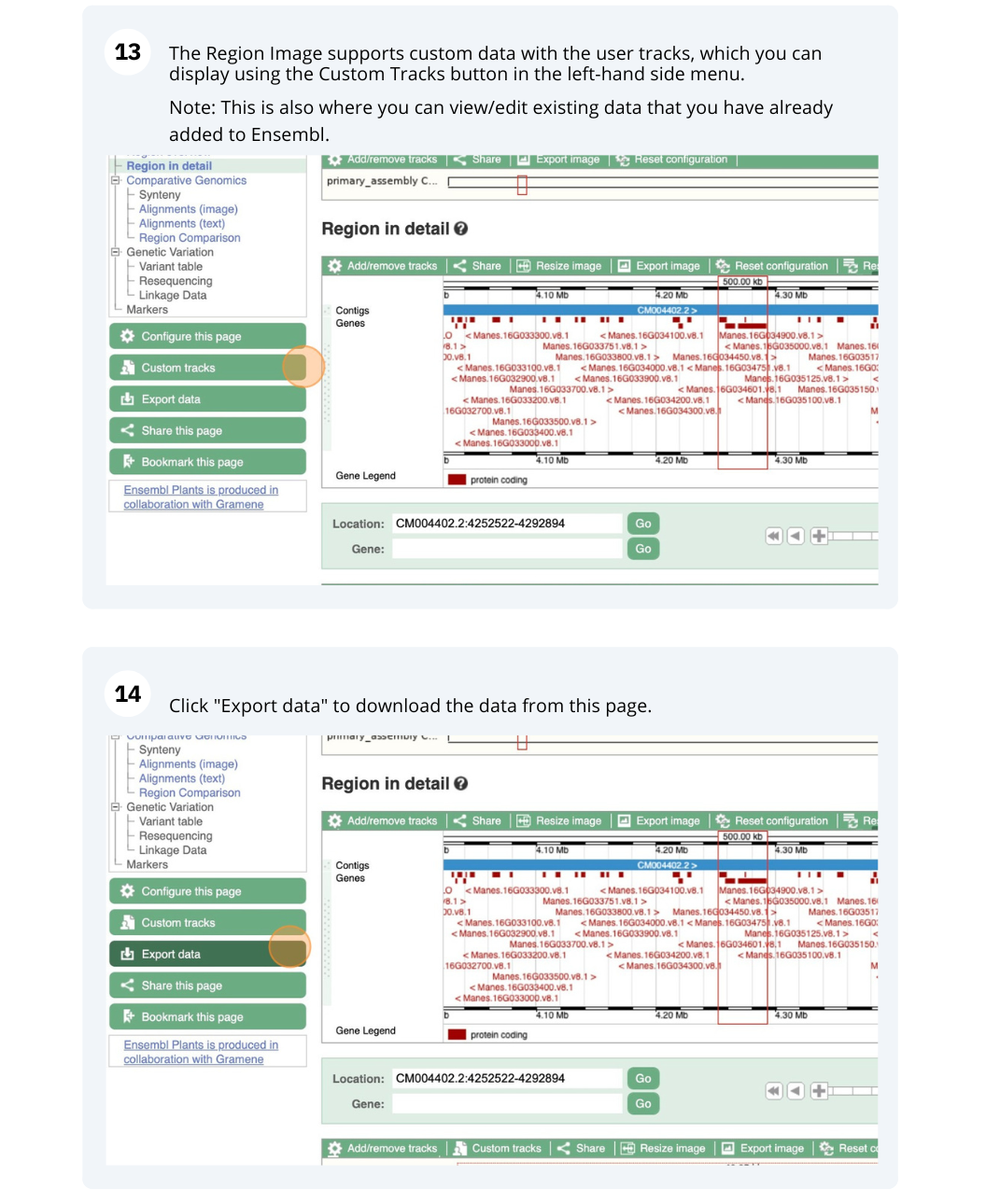
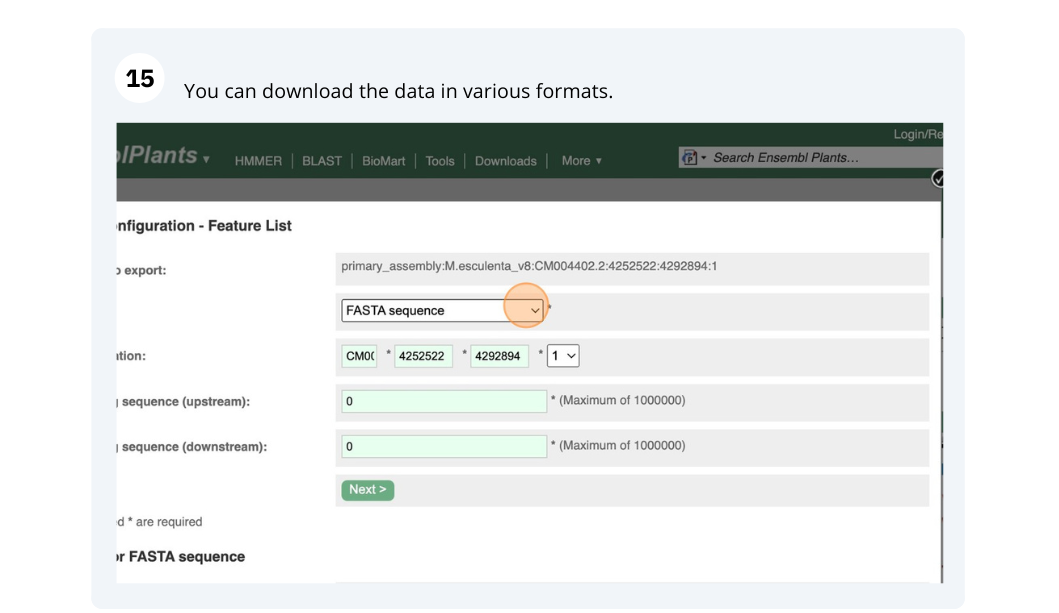
Exploring a genomic region in Oryza sativa Japonica (rice)
Go to the Ensembl Plants homepage and do the following:
-
Go to the region between 405000 and 453000 on chromosome 1 in Oryza sativa Japonica.
-
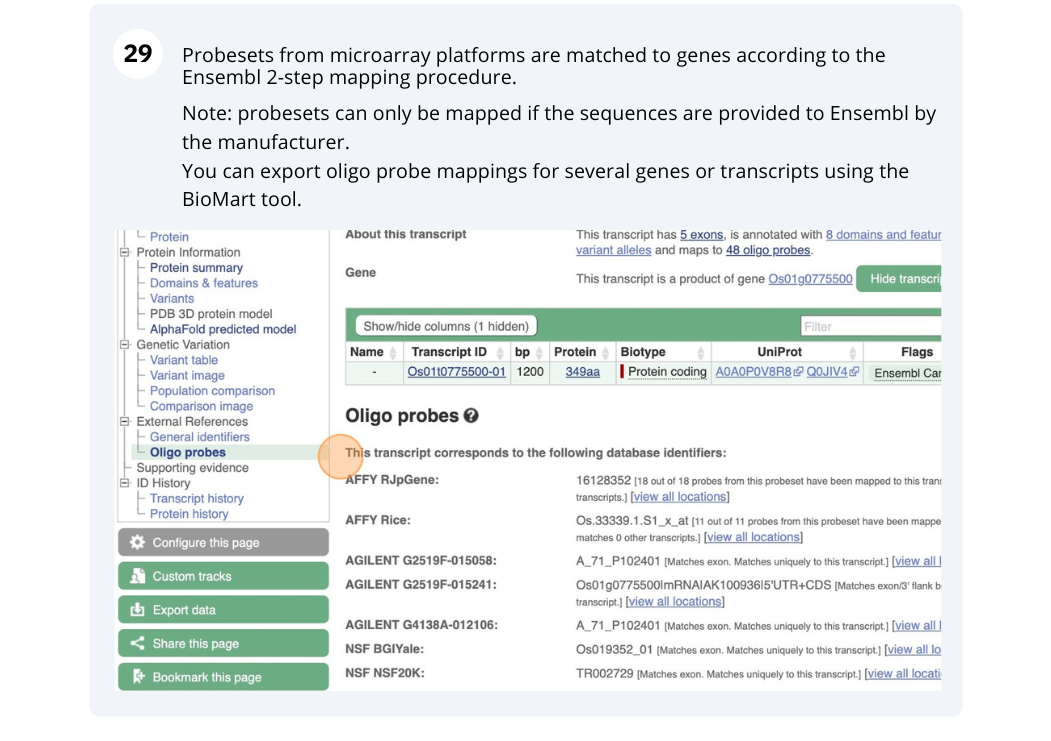
Turn on the AGILENT:G2519F-015241 microarray track. Are there any oligo probes that map to this region?
-
Highlight the region around any reverse strand probes you can see. Do they map to any Ensembl transcripts?
-
Go to the Ensembl Plants homepage. Select Oryza sativa Japonica from the Species drop-down list and type
1:405000-453000. Click Go. - Click on Configure this page to open the menu. You can find the AGILENT:G2519F-015241 track under Oligo probes in the left-hand menu, or by using the Find a track box at the top right. Turn on the track as Normal then save and close the menu. As the AGILENT:G2519F-015241 track is stranded, it appears at the top and bottom of the view.
There are 5 probes mapped to this region on the positive strand and one probe on the reverse strand.
- Drag a box around the reverse strand probe then click on Mark region to highlight.
The highlighted region maps to two transcripts: Os01t0107900-02 and Os01t0107900-01
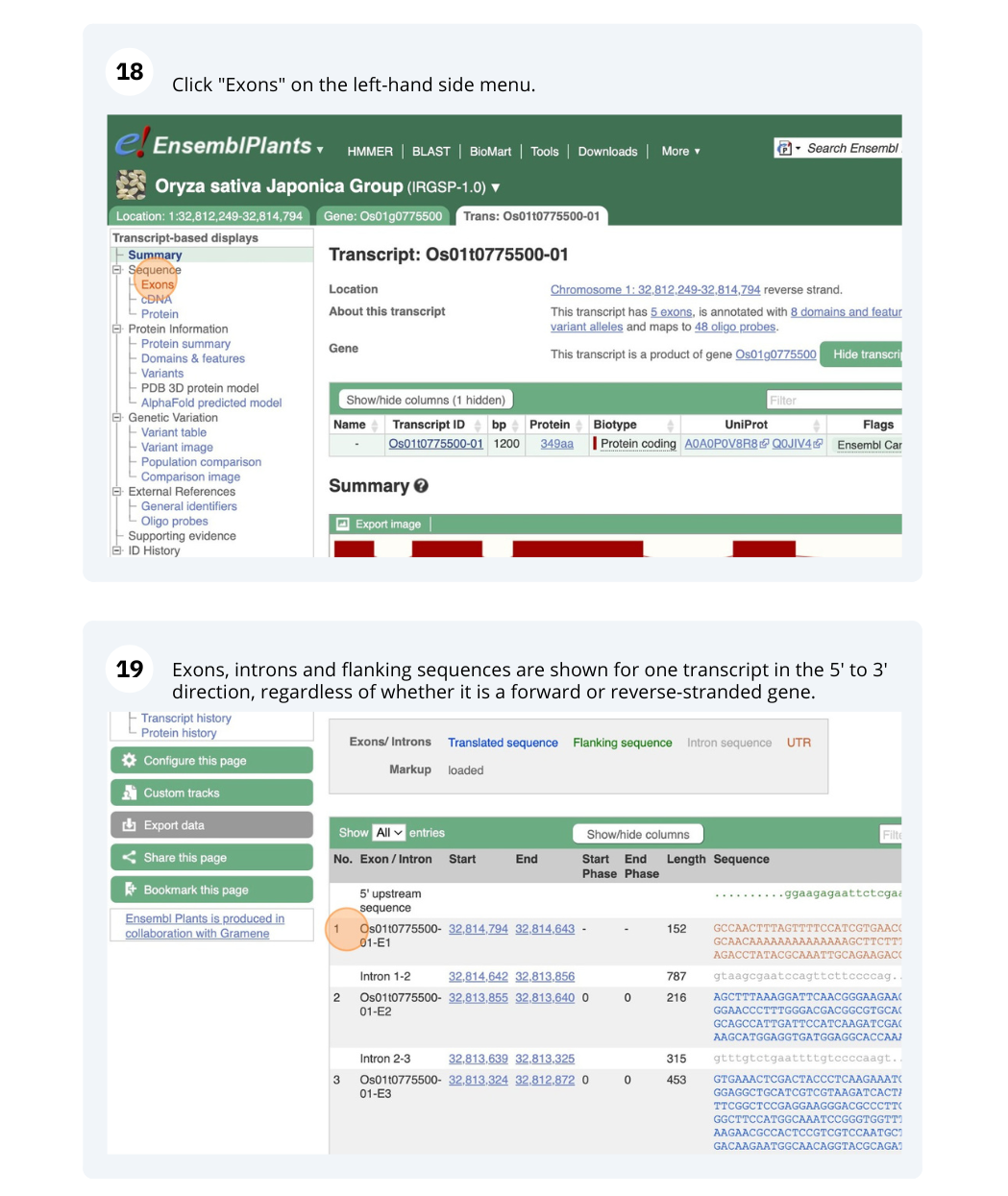
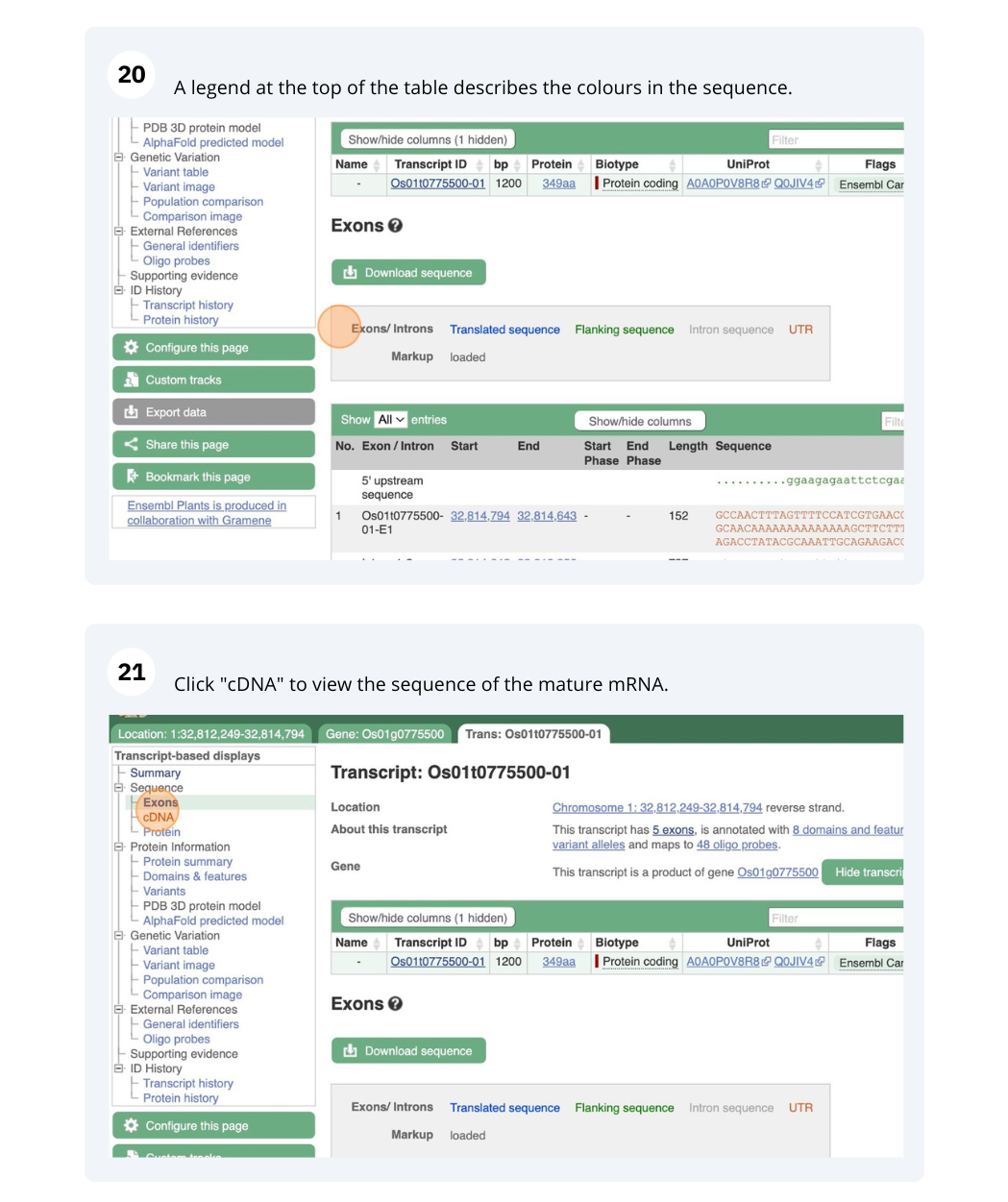
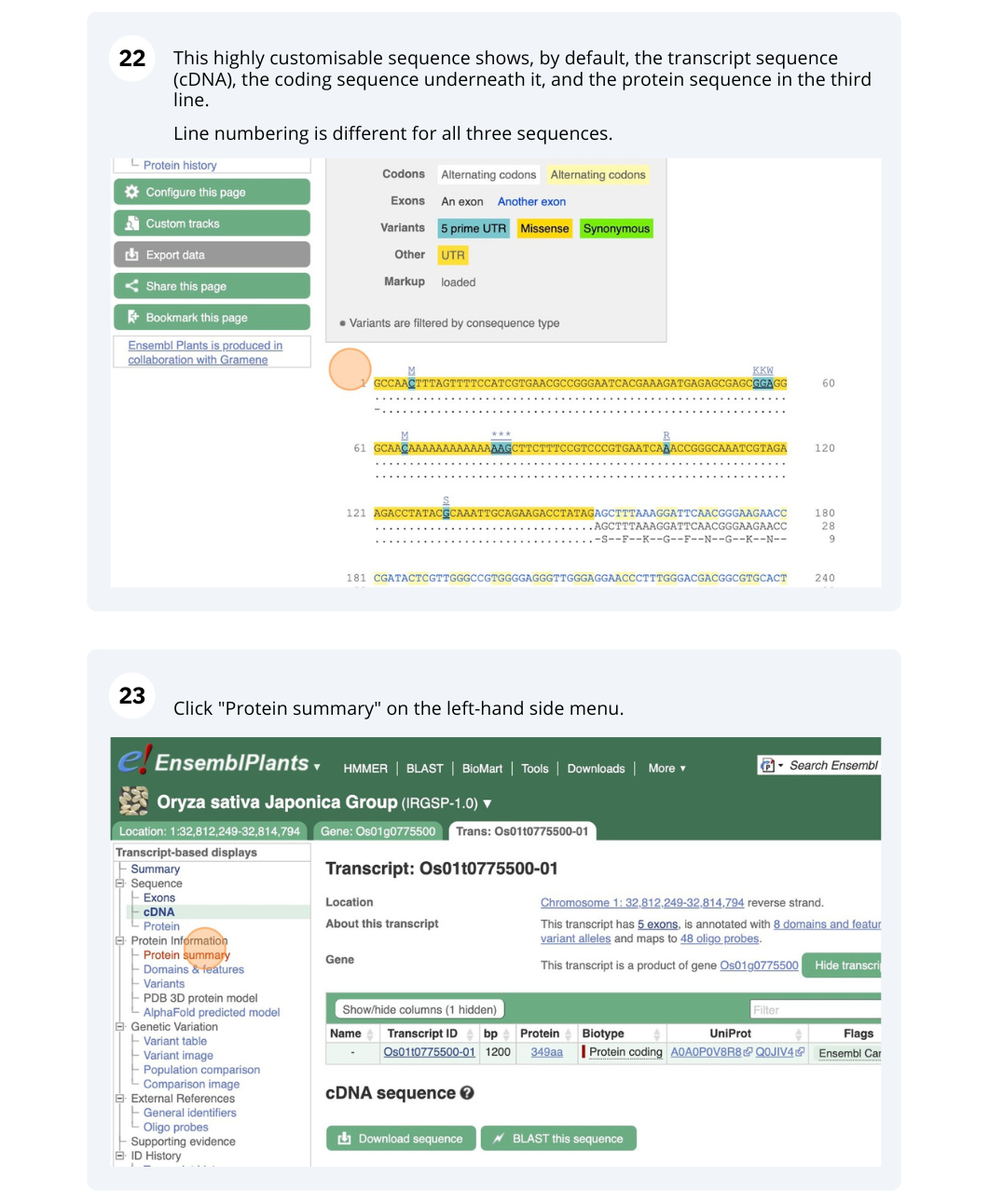
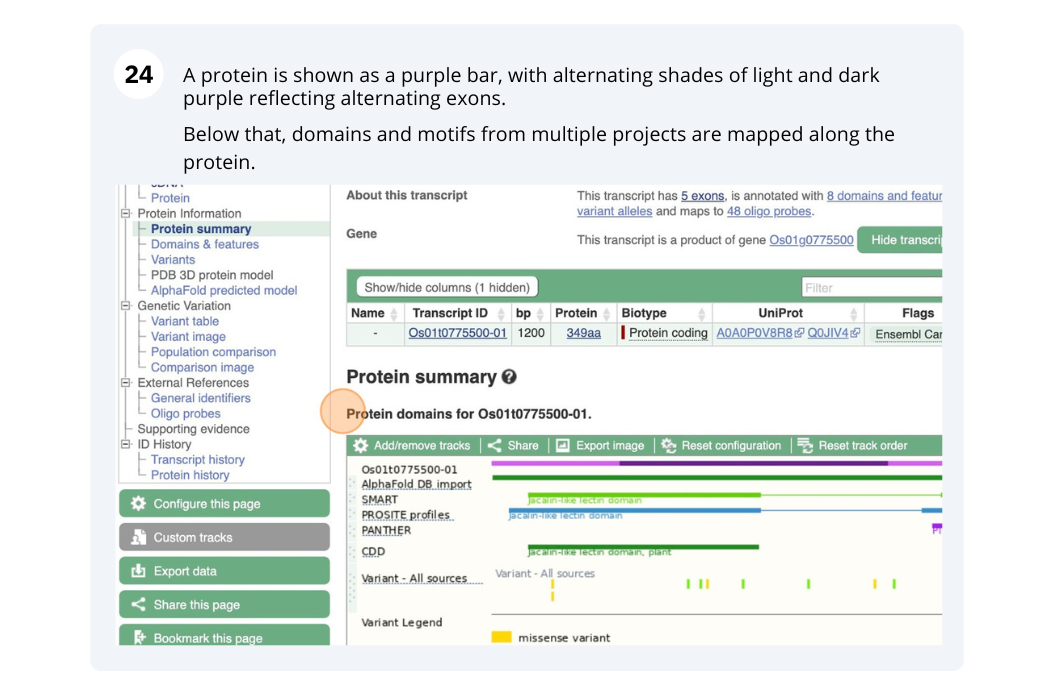
Genes and Transcripts
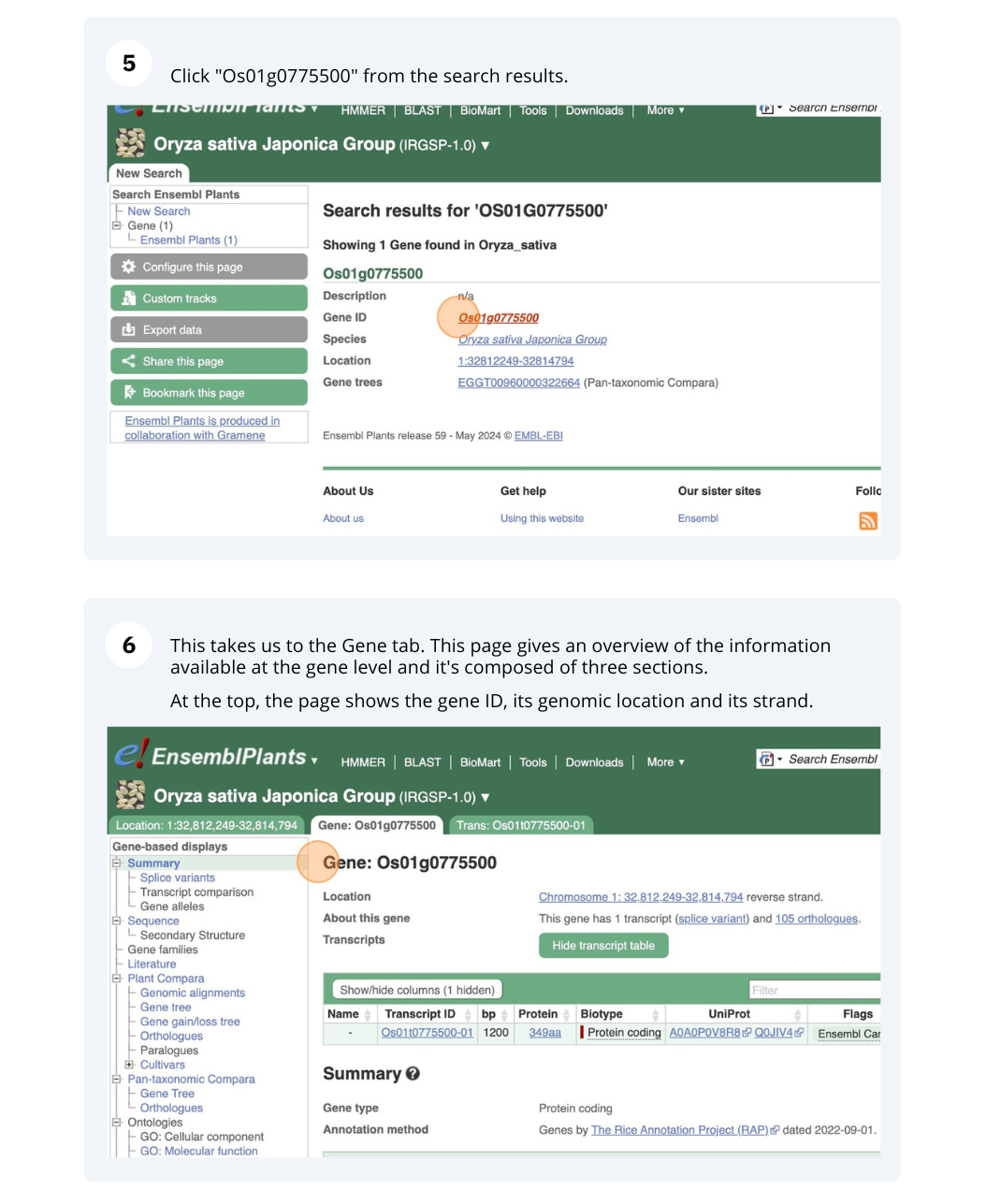
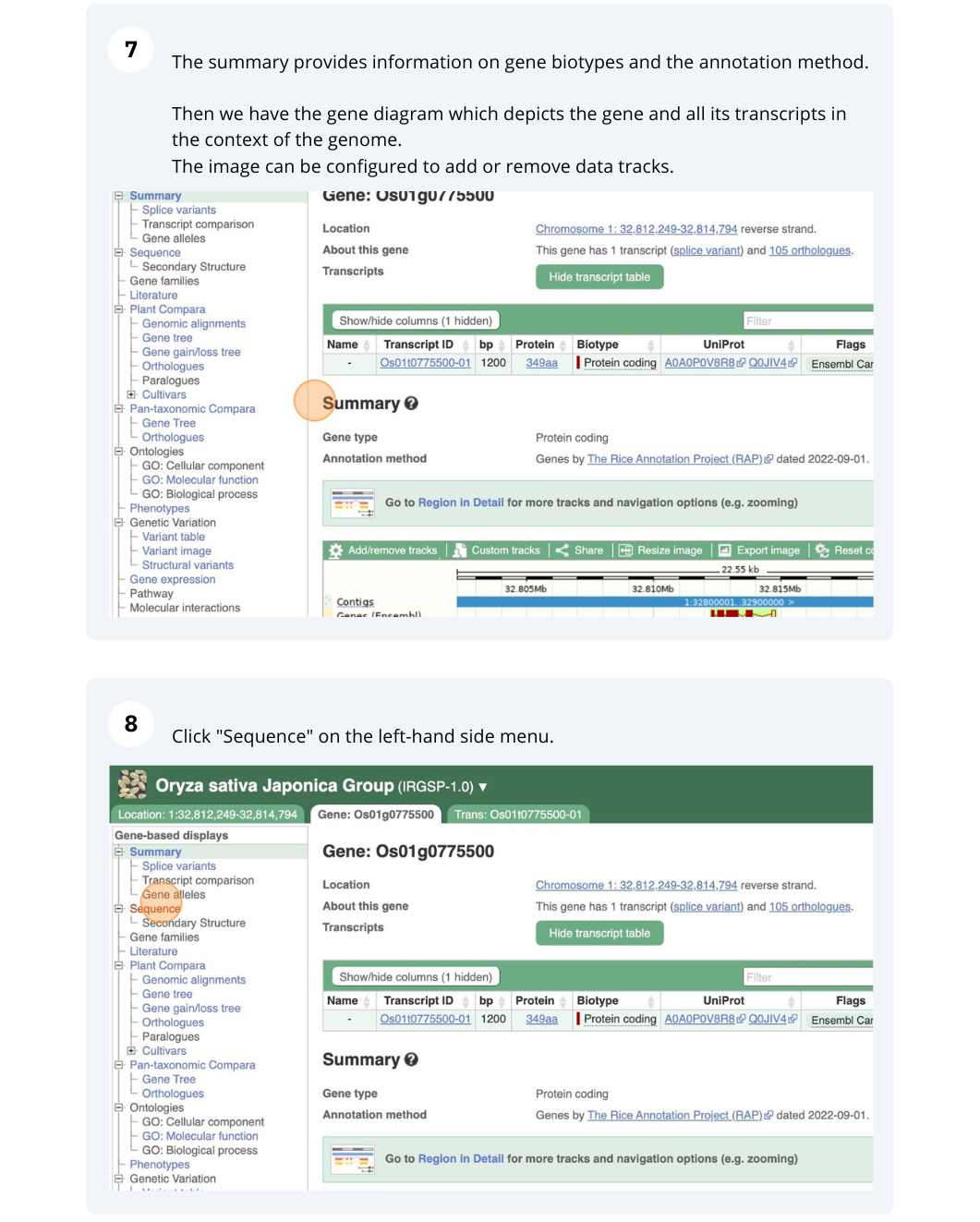
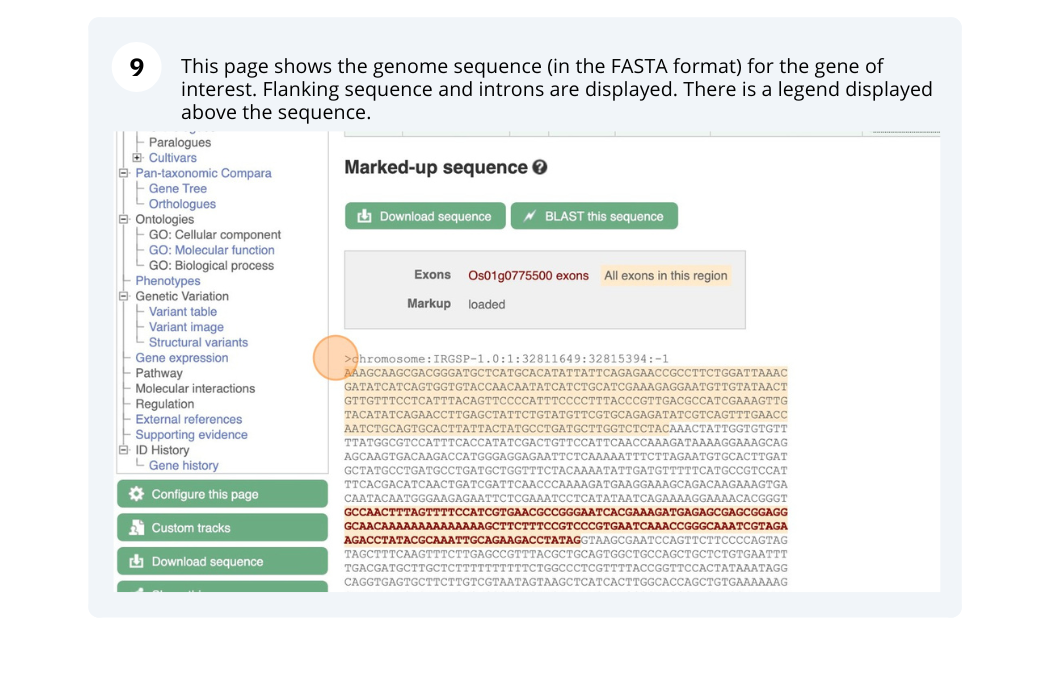
We will search for Oryza sativa Japonica Group (IRGSP-1.0) gene OS01G0775500 in Ensembl Plants.


Exploring a defence-related gene in Tomato, Solanum lycopersicum
(a) Search for the tomato gene NCED2 and go to the gene tab.
- What is the amino acid length of the only transcript of this gene?
- Which chromosome and which strand of the genome is this gene located?
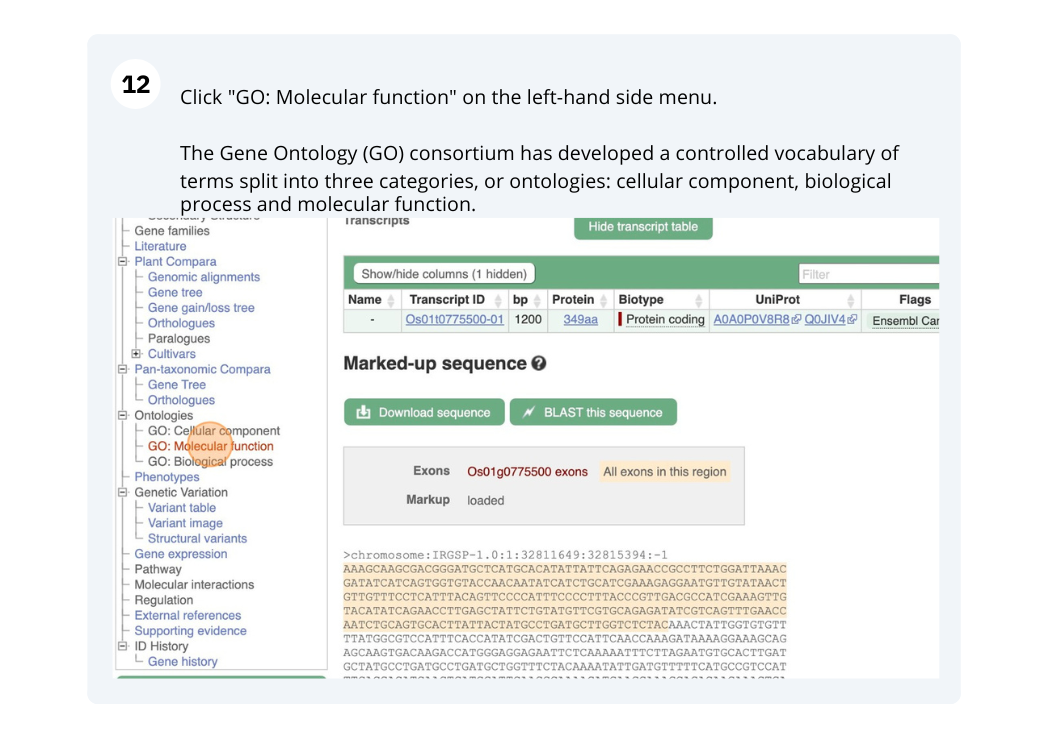
(b) Look at the gene Description field, what does this tell you about the cellular localisation of the protein product of this gene? Does this match the Gene Ontology (GO): Cellular component terms? Click on GO:Cellular component to check.
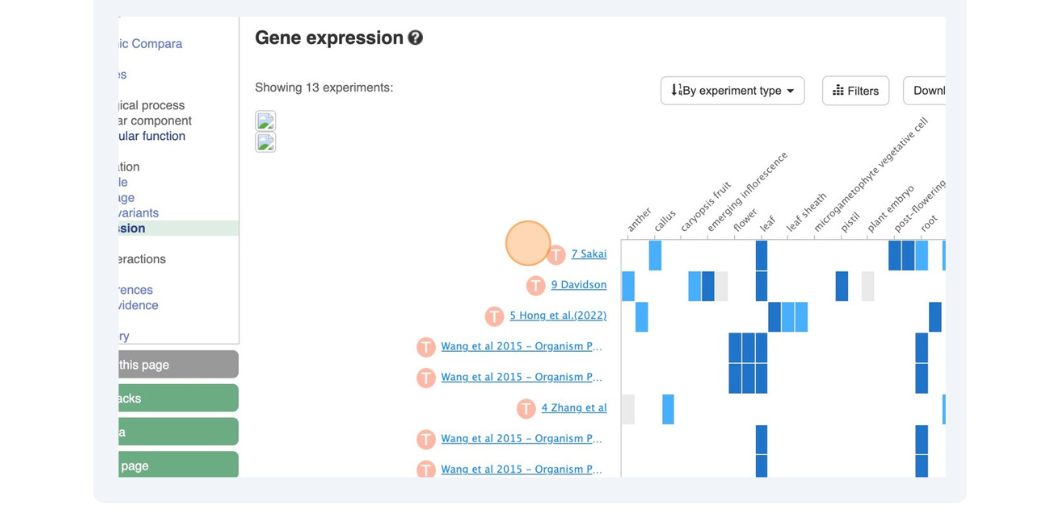
(c) Click on Gene expression. Which tissue has the highest expression of this gene according to the Tomato Genome Consortium?
(d) The summary at the top of the page (just above the Show transcript table button) shows us that there are nine paralogues of this gene. Click on the Gene gain/loss tree to look at the expansion of this gene family across all plants.
- Which species has the largest number of members of this gene family?
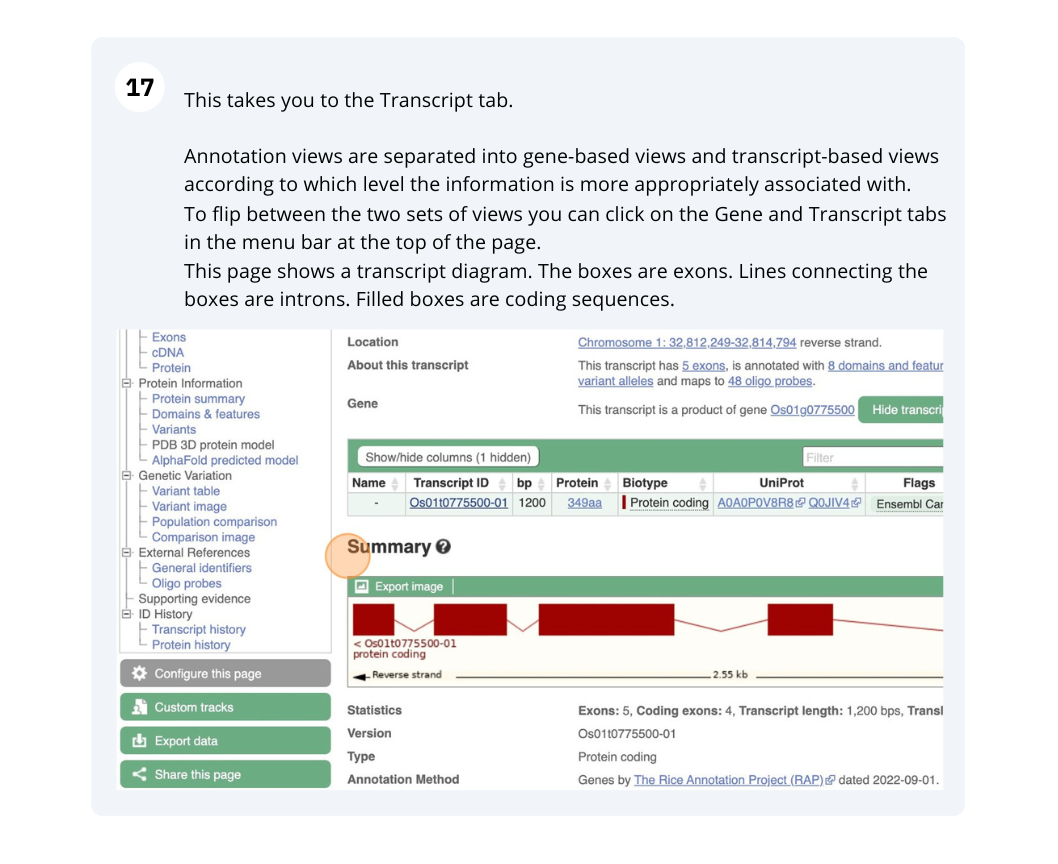
(e) Go to the transcript tab for this gene by clicking on the transcript ID Solyc08g016720.1.1 from the transcript table. Are there any Oligo probes that would be useful in targeting this gene experimentally?
(a) Go to plants.ensembl.org and type NCED2 into the search box, selecting Solanum locypersicum from the drop down menu. Click on the first result to go to the gene tab.
Click on the Show transcript table button if the transcript table is hidden. In the 4th column we see the protein length listed, 581 amino acids in length.
The location is listed at the top of the page, we can see that this is on Chromosome 8, between the base pairs 8,729,953 and 8,731,698, and on the forward strand.
(b) The gene description for this gene is ‘9-cis-epoxycarotenoid dioxygenase NCED2, chloroplastic’ which suggests the enzyme is localised to the chloroplast.
In the left-hand navigation panel, find the link to GO: Cellular location. We can see three results, chloroplast, plastid and chloroplast stroma, so this matches the gene description.
(c) Click on Gene expression in the left-hand navigation panel.
Darker shades of blue indicate higher expression. Hover your mouse over the heat-map to show a pop-up with the TPM (Transcripts Per Kilobase Million).
The 2cm fruit in the Tomato Genome Consortium has the highest expression at 103 TPM. You can also click on Filters at the top right and filter to high or medium expression.
(d) Click on the Gene gain/loss tree. You might find it easier to compare in the radial tree, click the two arrows icon at the top left of the image () to toggle to the radial view.
Look for the red lines, indicating the larger number of members and significant expansion. The number of members are listed just before the species name.
Brassica napus and Brassica juncea has the highest number of members in this gene family.
(e) Go to the transcript tab for this gene by clicking on the transcript ID Solyc08g016720.1.1 from the transcript table.
Find the Oligo probes link in the left-hand navigation panel. There is a single probe from Affymetrix, the AFFY TomGene, 20363698.
Exploring a fungal gene in Rosellinia necatrix
Rosellinia necatrix is is a fungal plant pathogen infecting several hosts including coffee, apples, apricots, avocados, cassava, strawberries, pears, hop, citruses and Narcissus, causing white root rot. A study by A. Zumaquero et al in 2019 (doi: 10.1186/s12864-019-6387-5) revealed SAMD00023353_4000440 as a gene potentially involved in pathogenesis.
Start in Ensembl Fungi and select the Rosellinia necatrix str. W97 (GCA_001445595) genome.
-
What GO: molecular function terms are associated with the SAMD00023353_4000440 gene?
-
Go to the transcript tab for the only transcript, GAP89412. How long is the transcript?
-
What domains can be found in the protein product of this transcript? How many different domain prediction methods agree with each of these domains?
- From the Ensembl Fungi homepage, select Rosellinia necatrix str. W97 (GCA_001445595) by selecting the species from the table of species. Type
SAMD00023353_4000440and click on the gene ID SAMD00023353_4000440. Click on GO: molecular function in the left-hand panel.There is one term listed: GO:0004190, aspartic-type endopeptidase activity.
- Click on the transcript named GAP89412 or on the Transcript tab.
GAP89412 is 1413 bp in length.
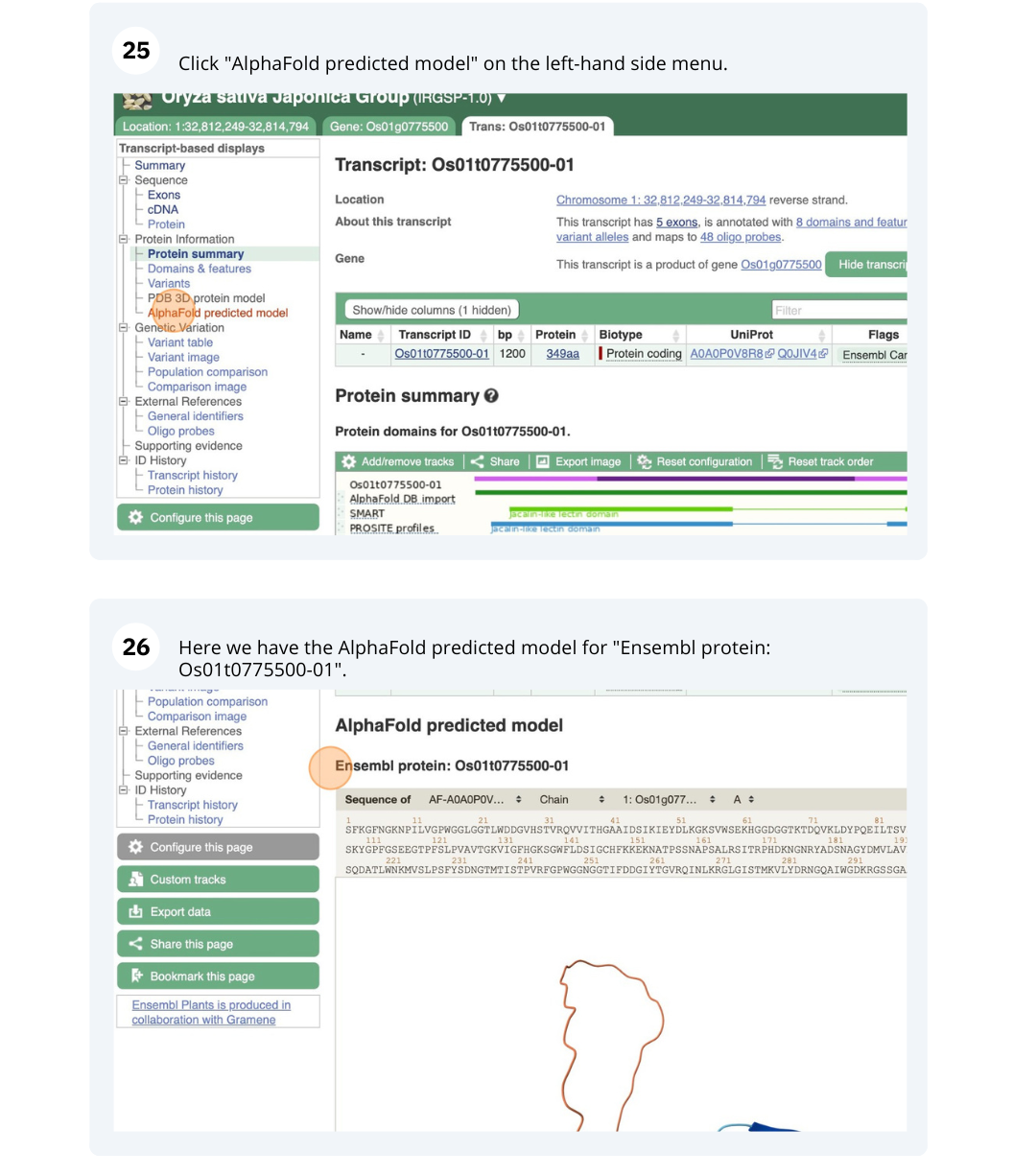
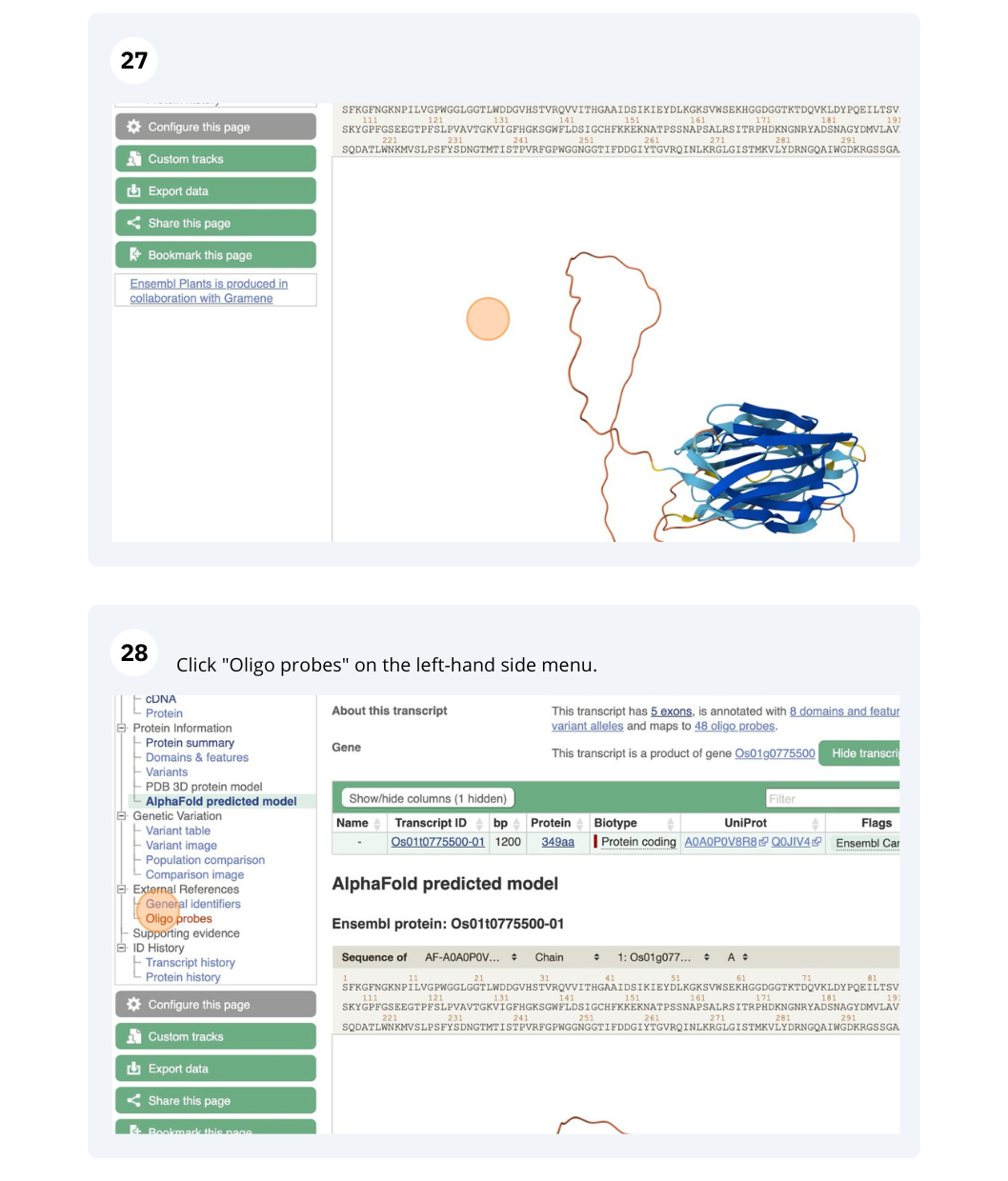
- Click on either Protein Summary or Domains & features in the left hand menu to see the predicted domains and motifs graphically or as a table respectively. You can also click on AlphaFold predicted model to view the AlphaFold predicted 3D structure of the protein.
Exploring a gene in Magnaporthe oryzae
We’re going to look at the gene ATG8 in Magnaporthe oryzae in Ensembl Fungi. This gene is involved in autophagy, and targeted silencing of this gene inhibits infection (you can find further info in Wilson and Talbot, Nature Reviews Microbiology volume 7, pages 185–195 (2009)).
-
Find the genomic sequence of the M. oryzae ATG8 gene. How many exons does the gene have?
-
Which biological processes annotated by Uniprot are associated with this gene?
-
How many transcripts does the gene have? Download the cDNA sequence in FASTA format to your computer.
-
Are there any entries of the gene in external databases? If so, which ones?
- From the Ensembl Fungi homepage, type ATG8 into the Search for a gene search bar, click the drop-down menu and select Magnaporthe oryzae and click the Go button. Click on the gene ID MGG_01062, which will open the Gene tab. Go to Sequence in the left-hand panel. ATG8 exons are in red font.
The ATG8 gene has 3 exons.
- Go to Ontologies: GO: Biological process in the left-hand panel.
The ATG8 gene has 2 Uniprot annotation GO terms associated with it: Autophagy and protein transport.
- Click on Show transcript table underneath the gene summary information at the top of the page.
The gene has 1 transcript (MGG_01062T0).
Click on Sequence: cDNA in the left-hand panel. You can export the sequence by clicking the Download sequence button, which will open a pop-up menu. Select FASTA from the drop-down list and select cDNA only. You can download the sequence as is or, if you have a large sequence, you can download the compressed file.
- Click on External References: General identifies in the left-hand panel. You will find hyperlinks to entries in external databases under the Database identifier column.
Yes, there are entries in the Magnaporthe comparative DB, NCBI gene and WikiGene.
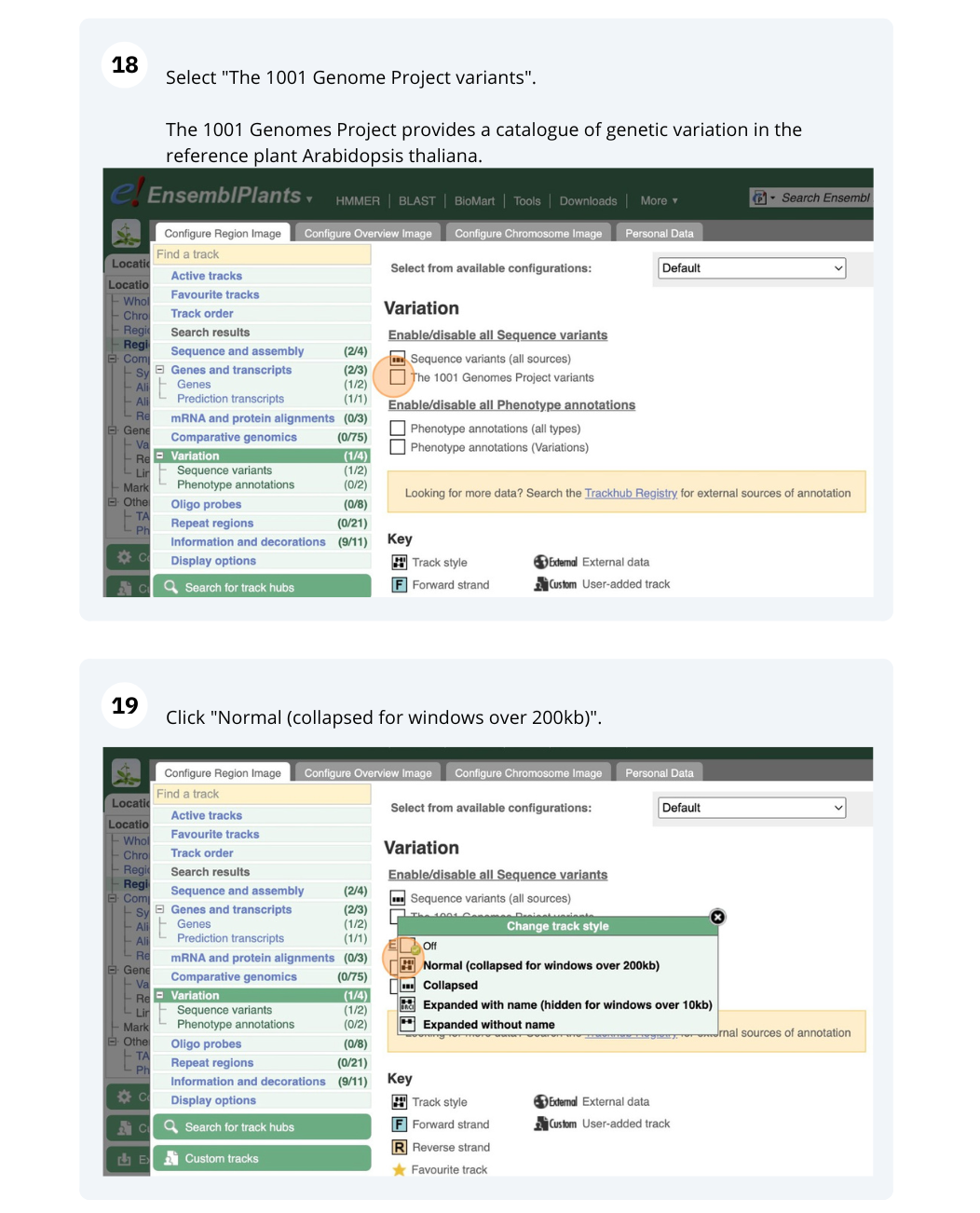
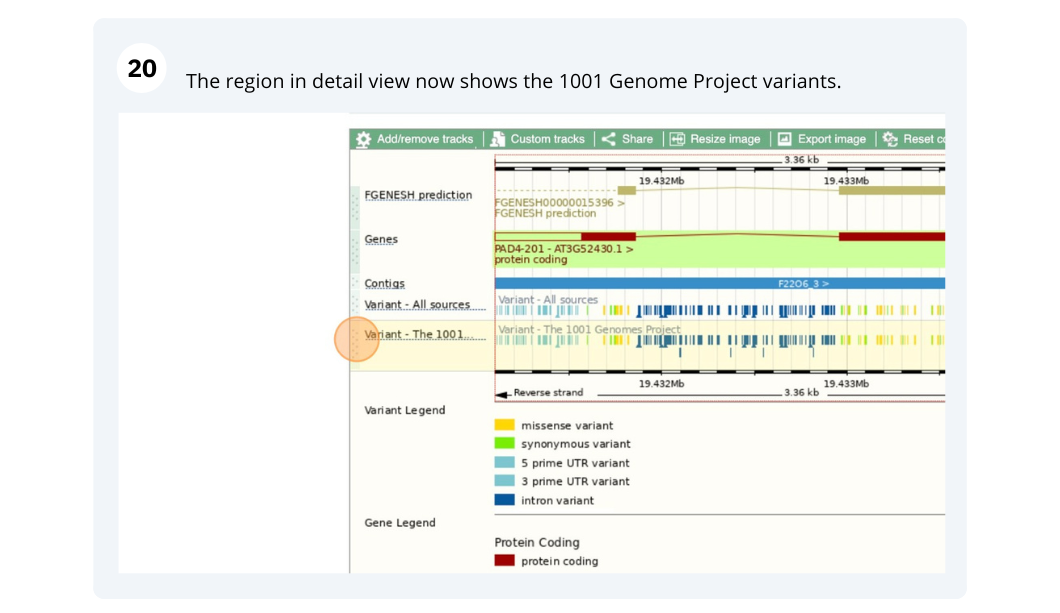
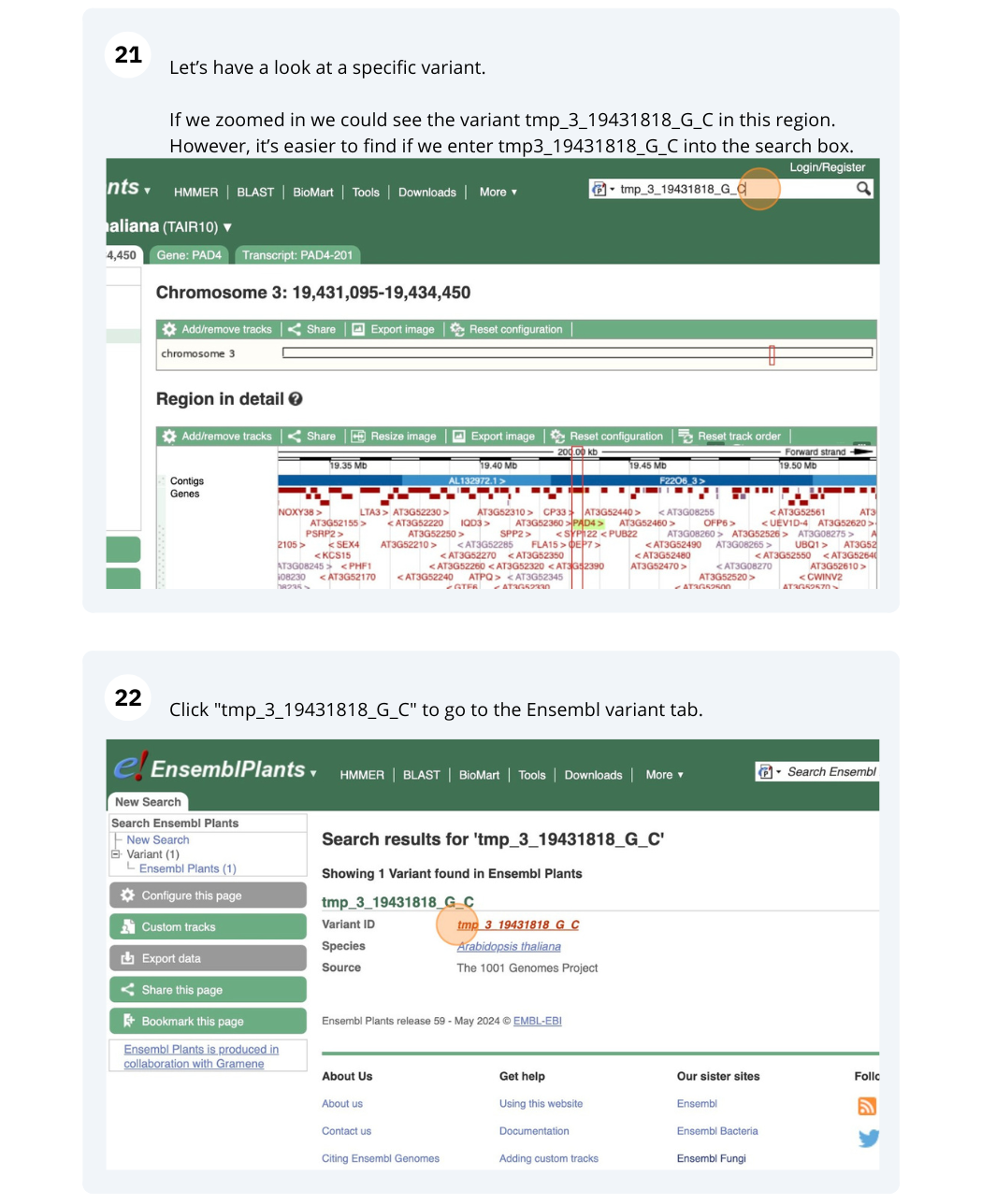
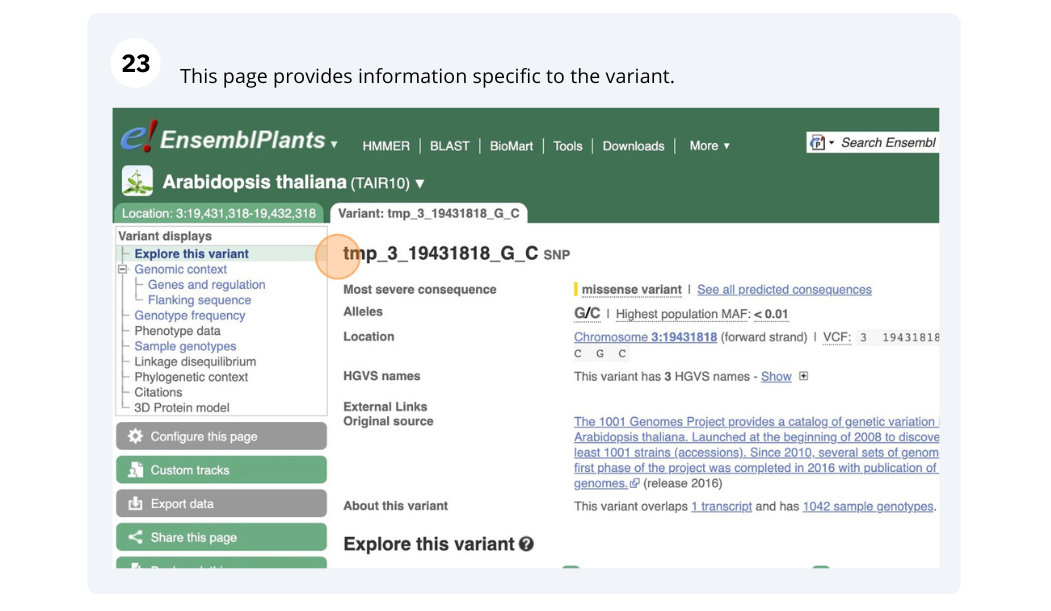
Variation
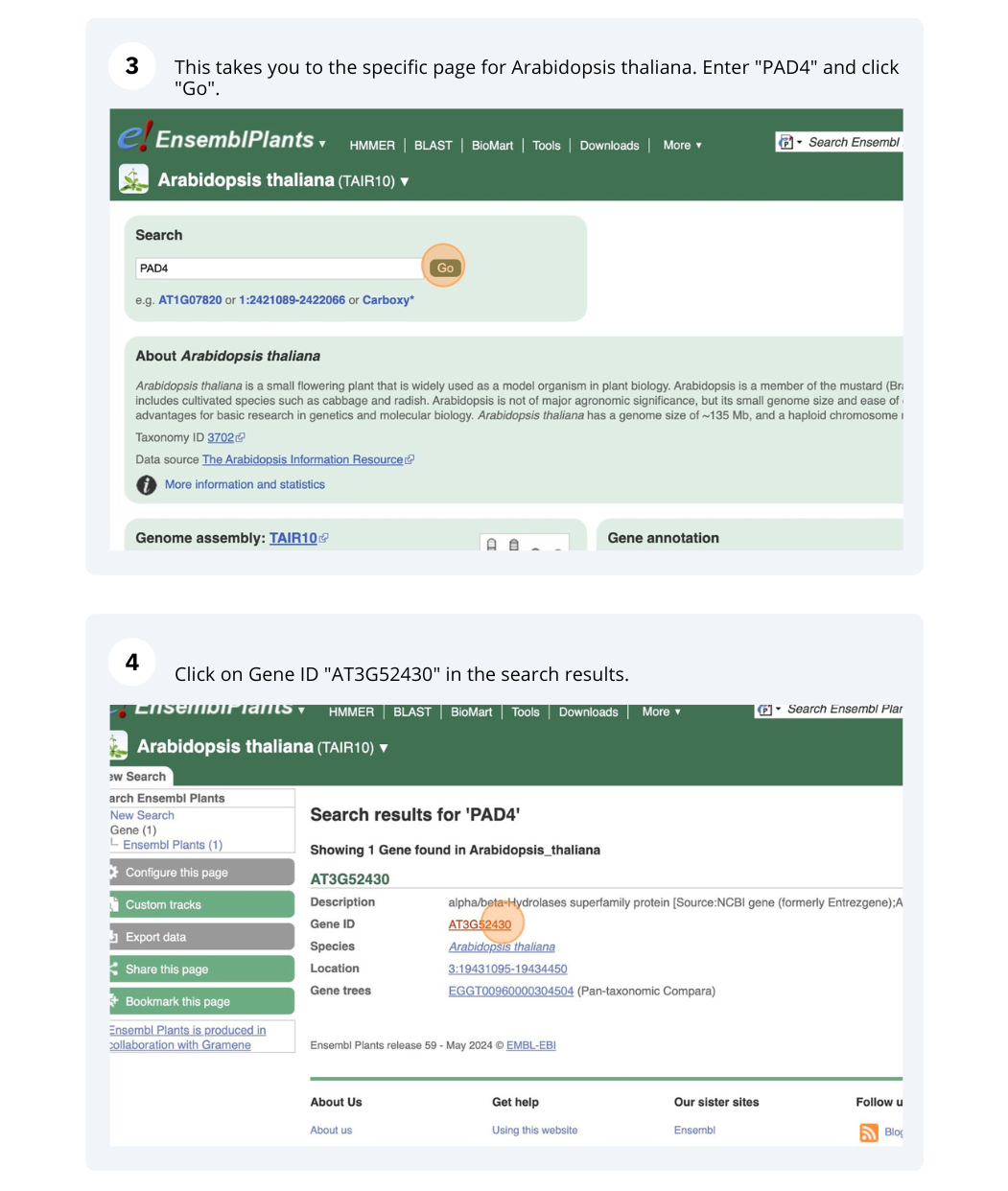
We are going to look at a gene PAD4 in Arabidopsis thaliana to find variants in the gene.
We will look at the region of PAD4 to find variants in the region.
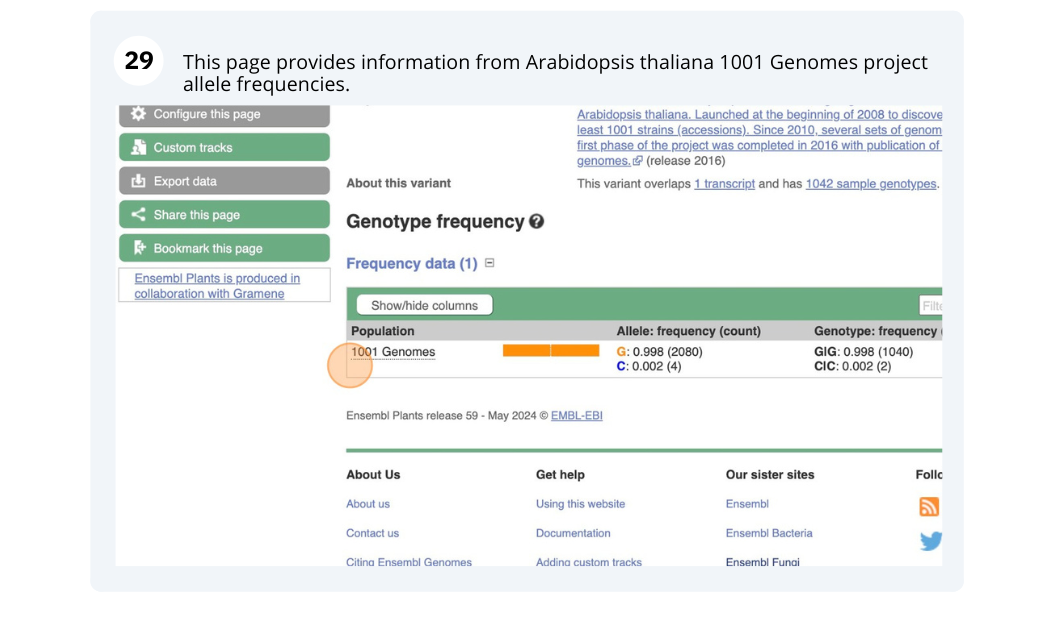
We will look at a variant tmp_3_19431818_G_C to find more information about it.
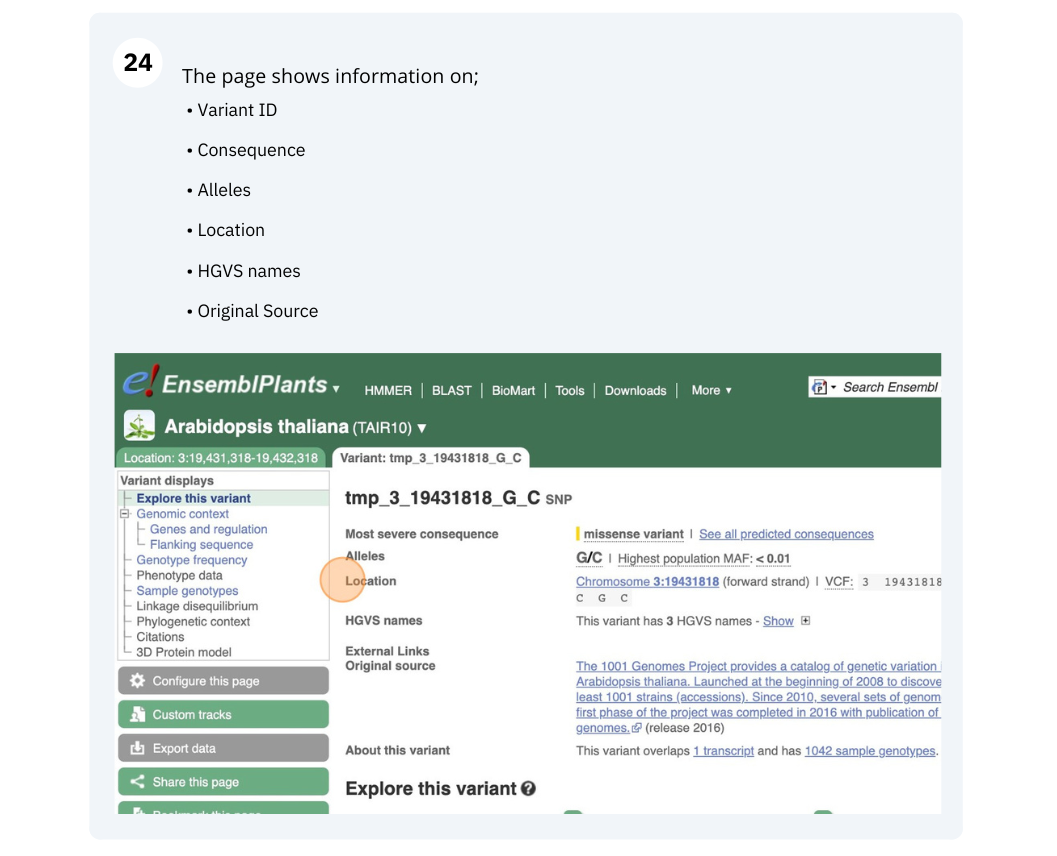
Exploring a SNP in Arabidopsis
The Arabidopsis thaliana ATCDSP32 protein is a chloroplastic drought-induced stress protein proposed to participate in a process called cell redox homeostasis. Go to Ensembl Plants and answer the following questions:
-
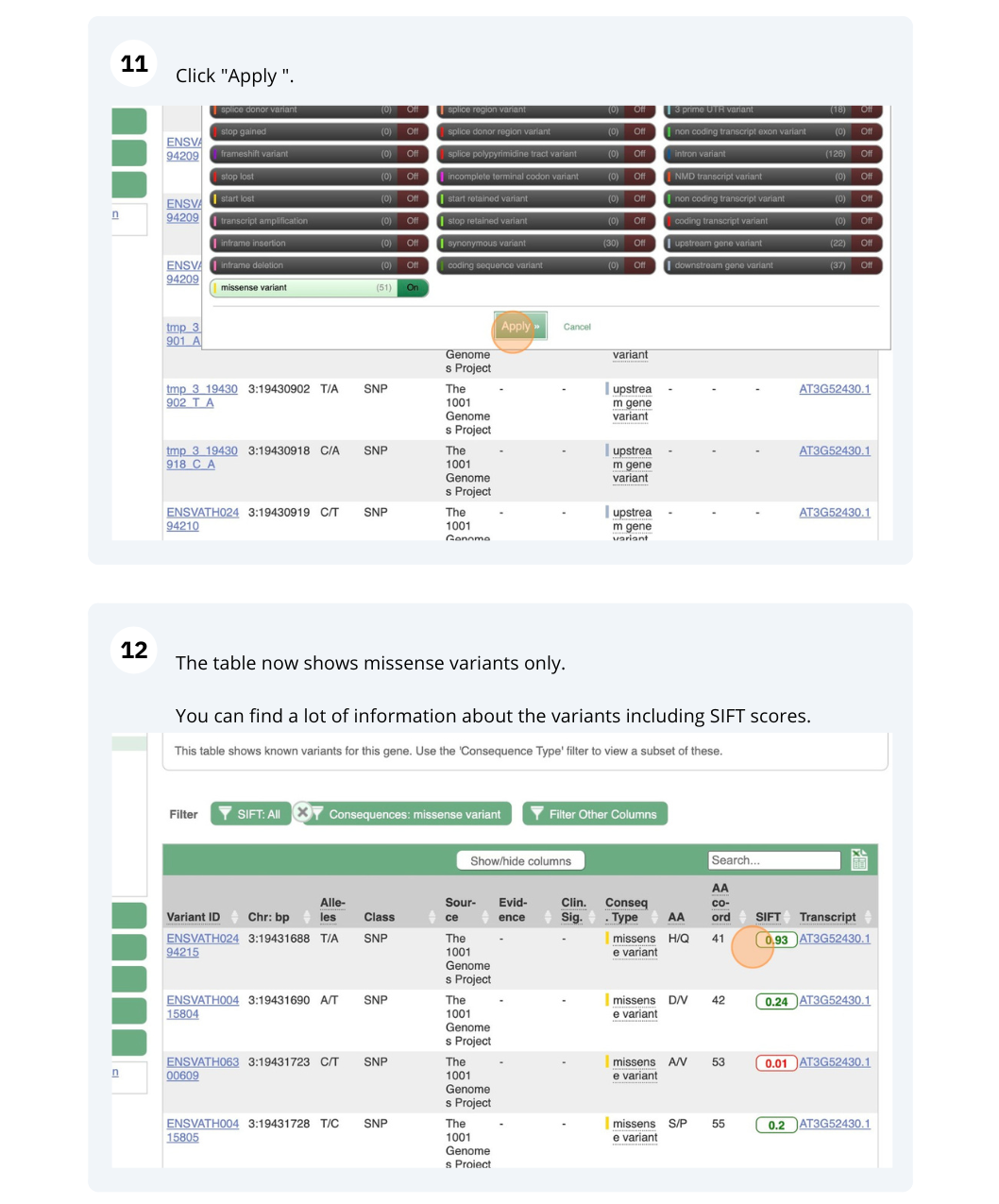
How many variants have been identified in the gene that can cause a change in the protein sequence (i.e. missense variant)?
-
What is the ID of the variant that changes the amino acid residue 60 from Alanine to Threonine (hint: refer to an amino acid codon table)? What is the location of this SNP in the A. thaliana genome? What are its possible alleles?
-
Download the flanking sequence of this SNP in RTF (Rich Text Format). Can you change how much flanking sequence is displayed on the browser?
-
Does this SNP cause a change at the amino acid level for other genes or transcripts?
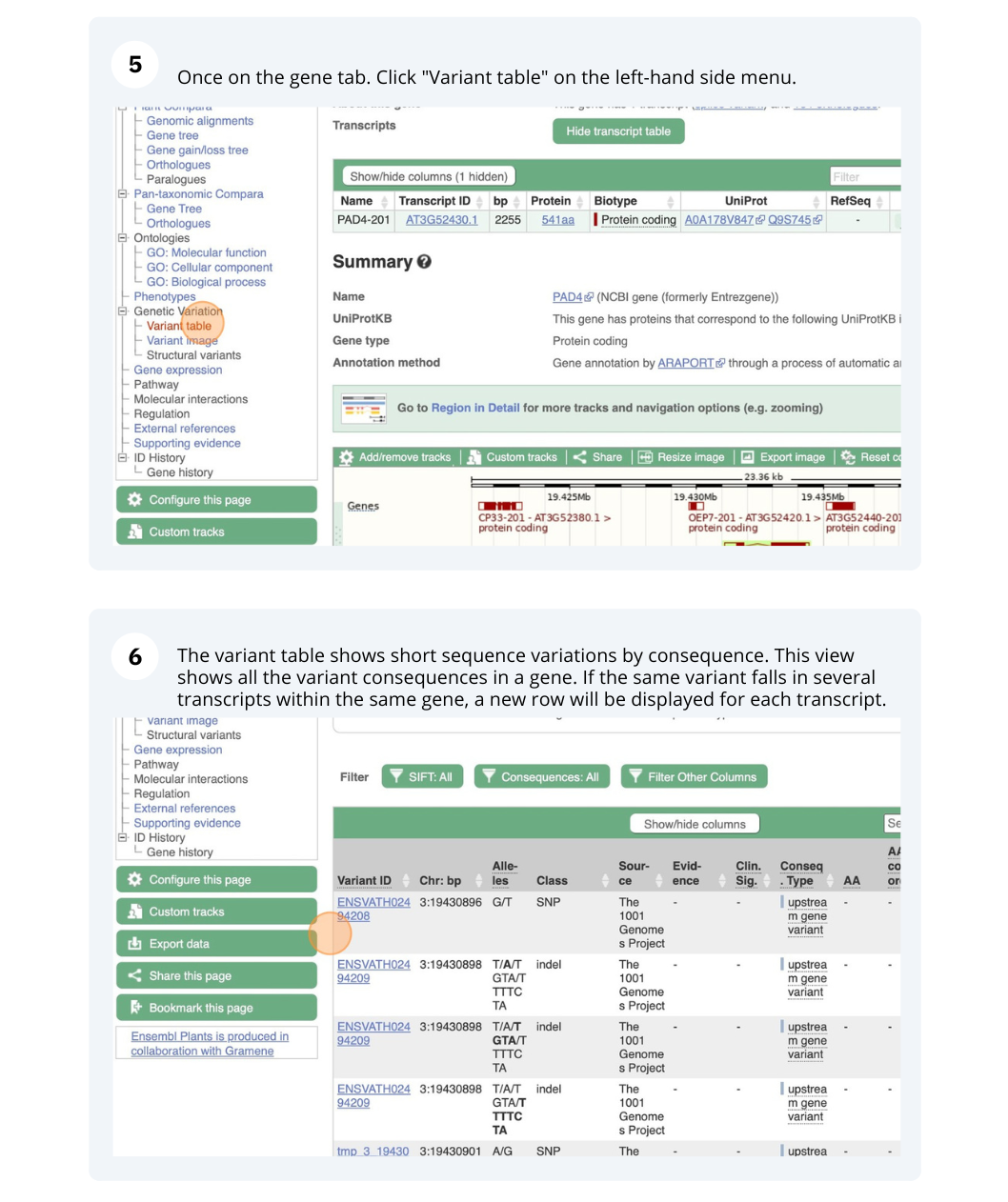
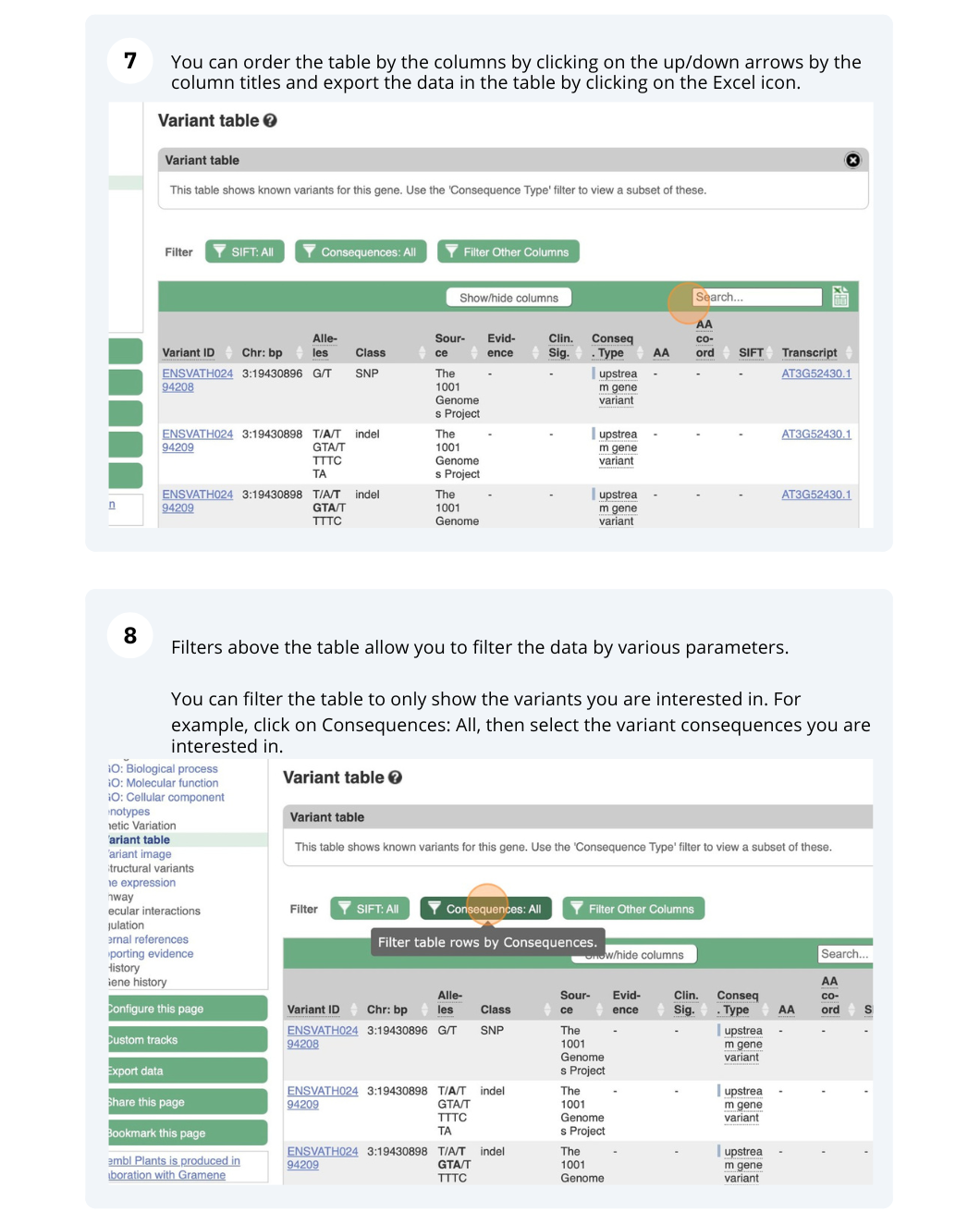
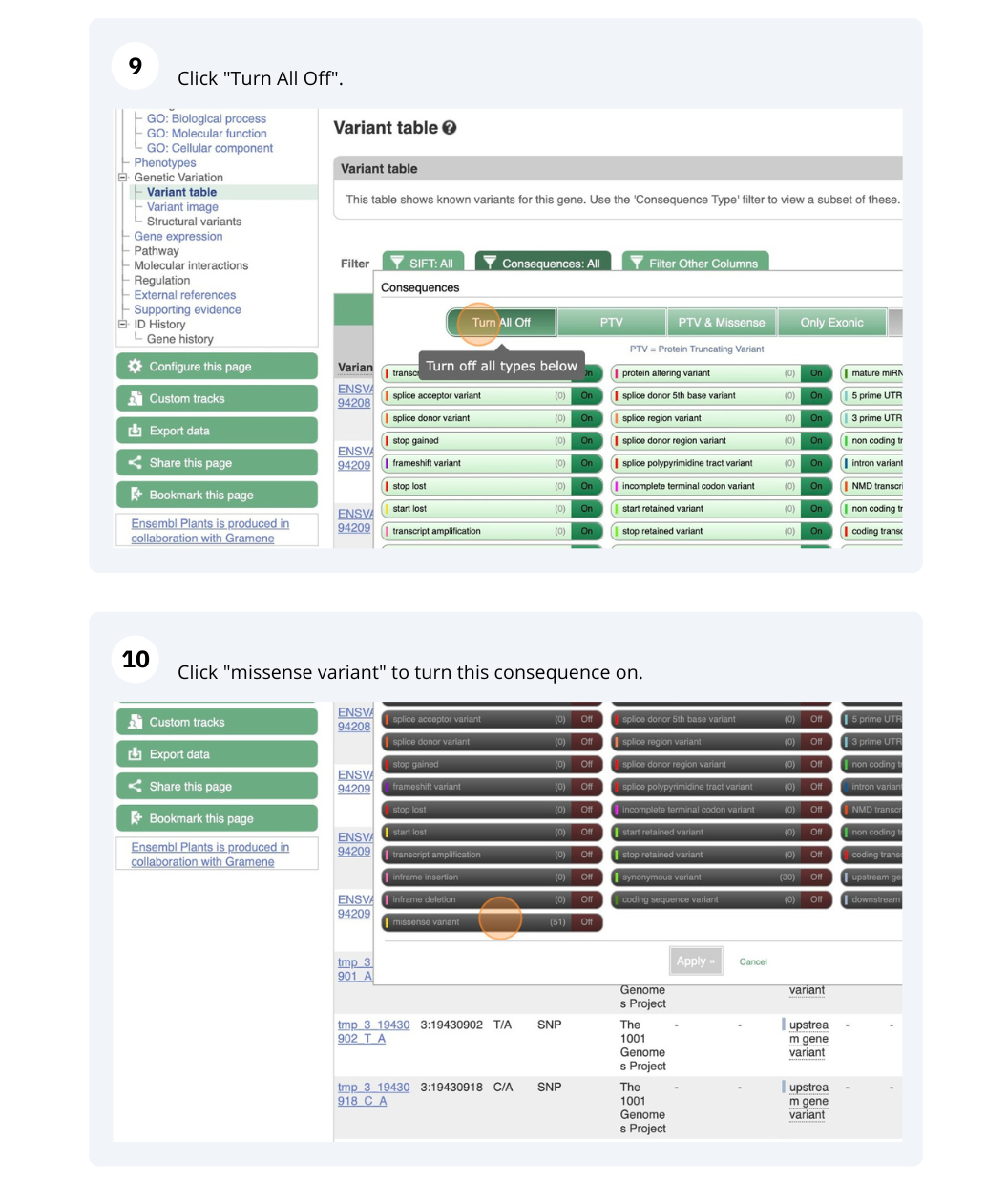
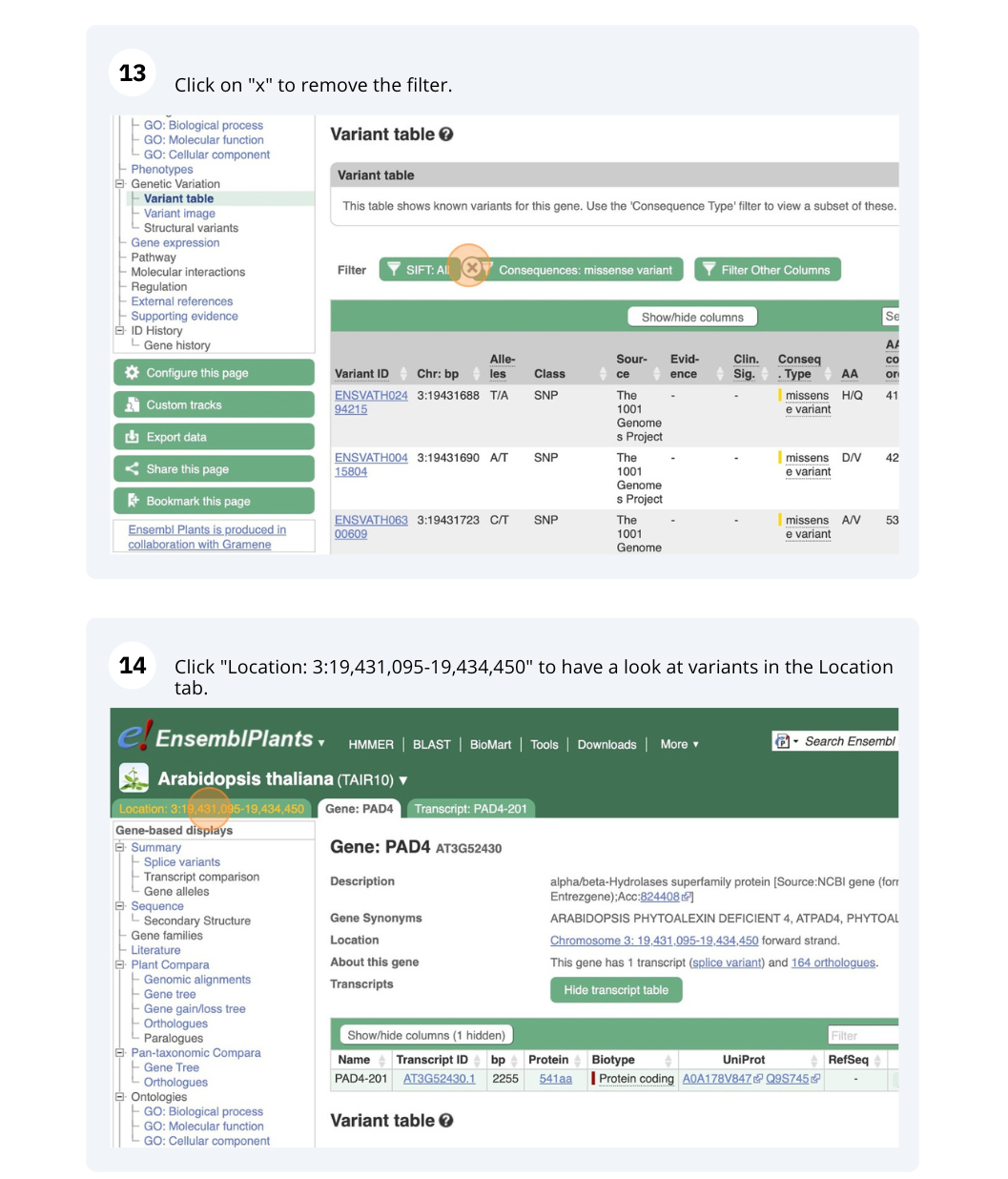
- Click on Arabidospsis thaliana on the Ensembl Plants homepage. Search for ATCDSP32 on the species page and in the search results, click on the Gene ID AT1G76080. In the left-hand side menu of the Gene tab, click on Variant table. Click on Consequences: All then select only missense variant.
The missense variant button indicates that there are 18 of these. Alternatively, you can count the number of variants in your filtered list.
- An amino acid codon table can be found on Wikipedia. Sort the AA coord column by clicking on the header and scroll down to find a variant at residue 60. The ID of this variant is ENSVATH05153232.
The variant is located at position 28549171 on chromosome 1. The two possible alleles at this locus are C (reference) and T (alternative).
- Click on the link ENSVATH05153232, then click on Flanking sequence in the left-hand side menu. Now click on Download sequence and select File format > Rich Text Format (RTF).
If you want to change how much flanking sequence is displayed on the browser, go back to the Flanking sequence page, click on the Configuration button and change the length of the sequence. The default settings is 400 bp.
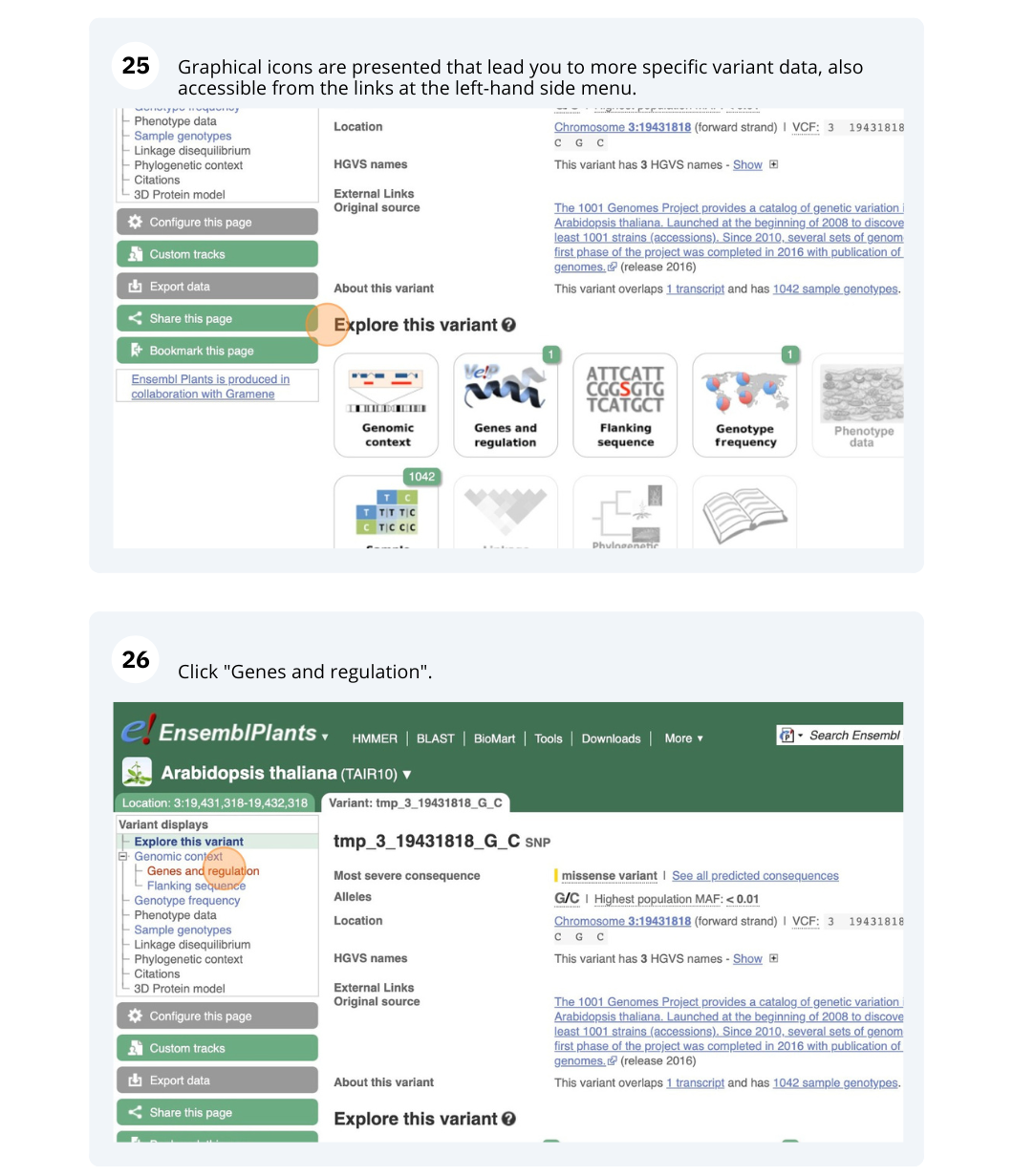
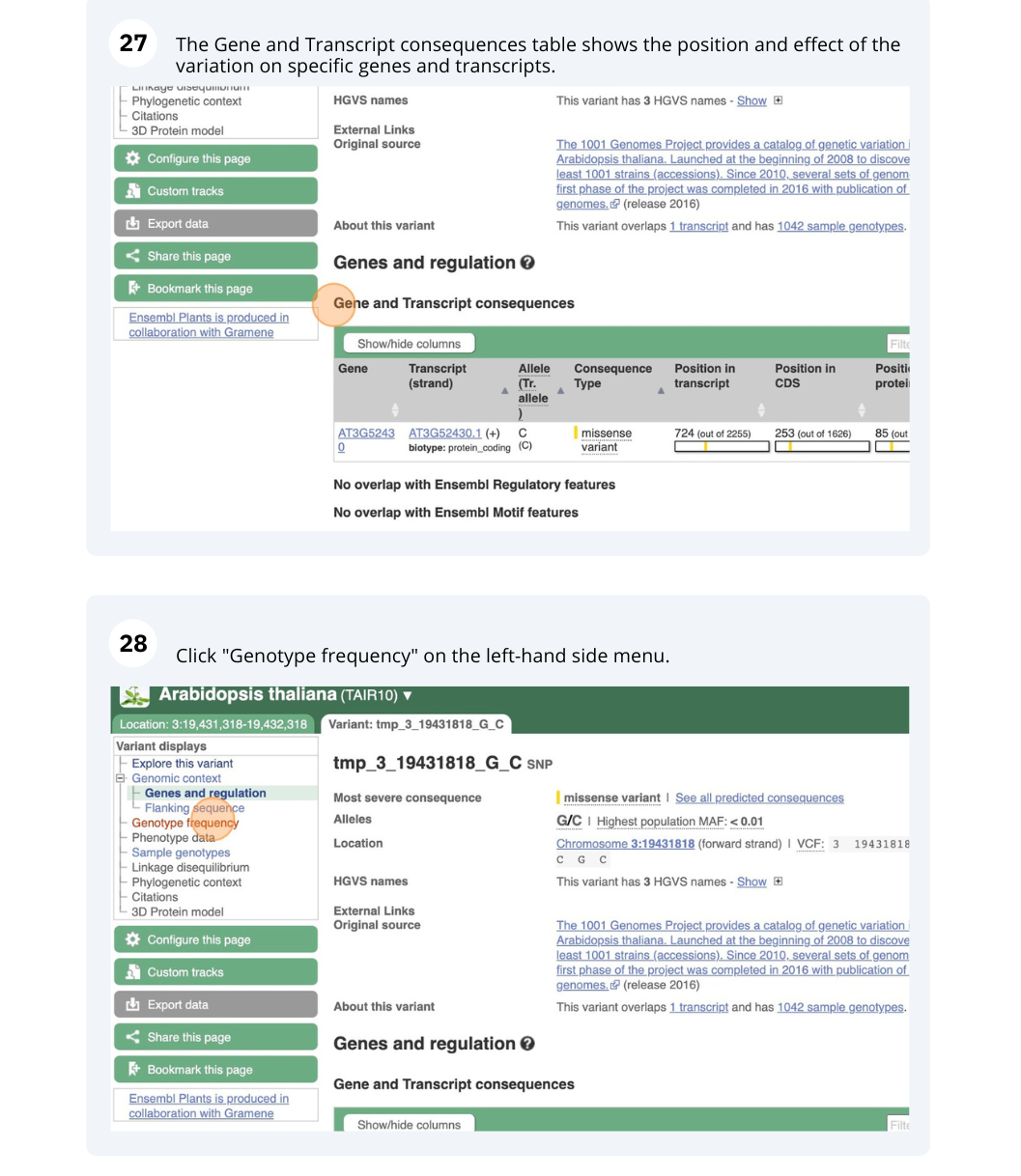
- Click on Genes and regulation in the left-hand side menu.
This SNP does not cause a change at the amino acid level for any other genes or transcripts in A. thaliana.
Variation data in Phaseolus vulgaris
-
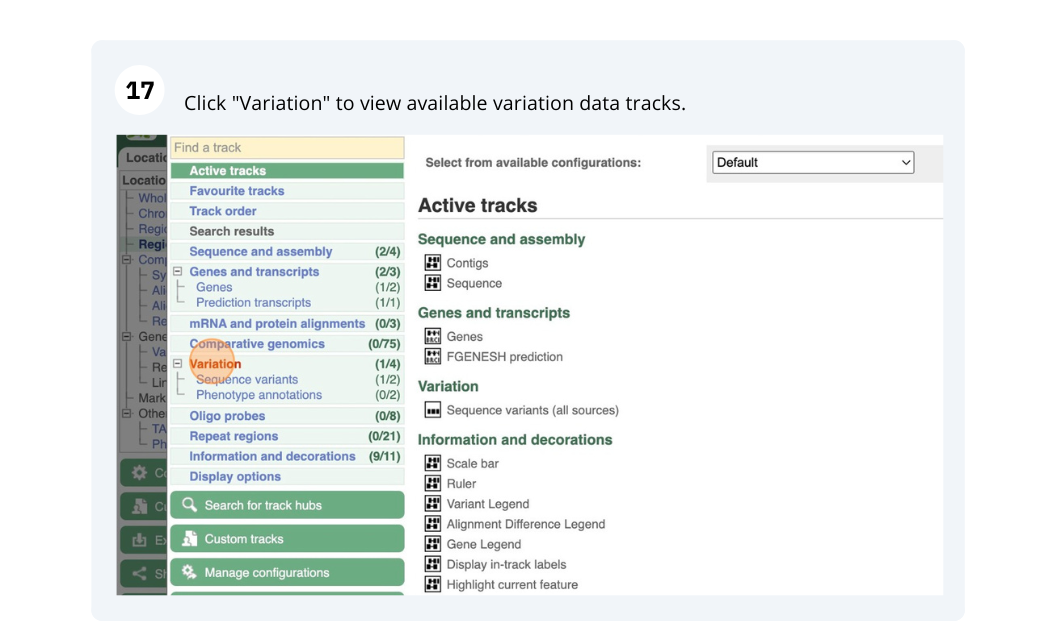
Go to Ensembl Plants and find the PHAVU_001G219900g gene in Phaseolus vulgaris and go to its Location tab. Can you see the variation track?
-
Zoom in around the first exon of this gene. Are any missense variants mapped in the translated region of this exon?
- Select Phaseolus vulgaris from the Species search drop-down menu and search for
PHAVU_001G219900g. In the results page, you can click on the coordinates 1:48,238,848-48,245,168 to go straight to the Location tab. Scroll down to the Region in detail view. The variation track (phaseolus_vulgaris_eva_PRJEB18671) is shown at the bottom of the view.If you don’t see the Variation - All sources track, click Configure this page on the left-hand panel, search for the track in the pop-up menu and enable the track by clicking on the square next to the track name. Close the pop-up window and wait for the track to load.
- Zoom in around the first exon of this gene by drawing a box in the respective region (you can change your mouse action by clicking the Drag/Select icons at the top right-hand corner of the view). Note the gene is on the reverse strand (this is signified by the < sign next to the transcript name, and it is located below the Contigs track), so the first exon will be on the right hand side of that image. The variation legend is shown at the bottom of the page, telling you what the colours mean.
There are four missense variants within the region; 1:48244305:C_T:PRJEB18671, 1:48244362:T_G:PRJEB18671, 1:48244426:G_A:PRJEB18671, 1:48244435:T_A:PRJEB18671.
Missense variants are shown in yellow. Click on the variants to get additional information on that variant including location. You can zoom into the region if the variant block is too small to click.
The variants are found at 1:48244305, 1:48244362, 1:48244426, 1:48244435.mSNPs are tagged with ambiguity codes (zoom into the region if you cannot see this). You can find a useful IUPAC ambiguity code guid on the bioinformatics.org website.
Variation data in tomato
-
Go to Ensembl Plants and find the Solyc02g084570.3 gene in Solanum lycopersicum (tomato) and go to its Location tab. Can you see the variation track?
-
Zoom in around the last exon of this gene. What are the different types of variants seen in that region? Are any splice region variants mapped in the region? If so, what is/are the coordinate(s)?
- Select Solanum lycopersicum from the Species search drop-down menu and search for
Solyc02g084570.3. In the results page, you can click on the coordinates 2:48284598-48288482 to go straight to the Location tab. Scroll down to the Region in detail view. The variation track is shown at the bottom of the view.If you don’t see the Variation - All sources track, click Configure this page on the left-hand panel, search for the track in the pop-up menu and enable the track by clicking on the square next to the track name. Close the pop-up window and wait for the track to load.
- Zoom in around the last exon of this gene by drawing a box in the respective region (you can change your mouse action by clicking the Drag/Select icons at the top right-hand corner of the view). Note the gene is on the reverse strand (this is signified by the < sign next to the transcript name, and it is located below the Contigs track), so the last exon will be on the left hand side of that image. The variation legend is shown at the bottom of the page, telling you what the colours mean.
The types of variants seen in that region are 3’ UTR, missense, synonymous and splice region variants.
Splice region variants are shown in orange. Click on the variants to get additional information on that variant including location. You can zoom into the region if the variant block is too small to click.
The variants are found at 2:48285642 and 2:48285640-48285641. Note that the two variants overlap: one is a SNP and the other is an indel. SNPs are tagged with ambiguity codes (zoom into the region if you cannot see this). You can find a useful IUPAC ambiguity code guid on the bioinformatics.org website. Single-letter ambiguity codes are given when two or more possible nucleotides may be represented at a single base locus.
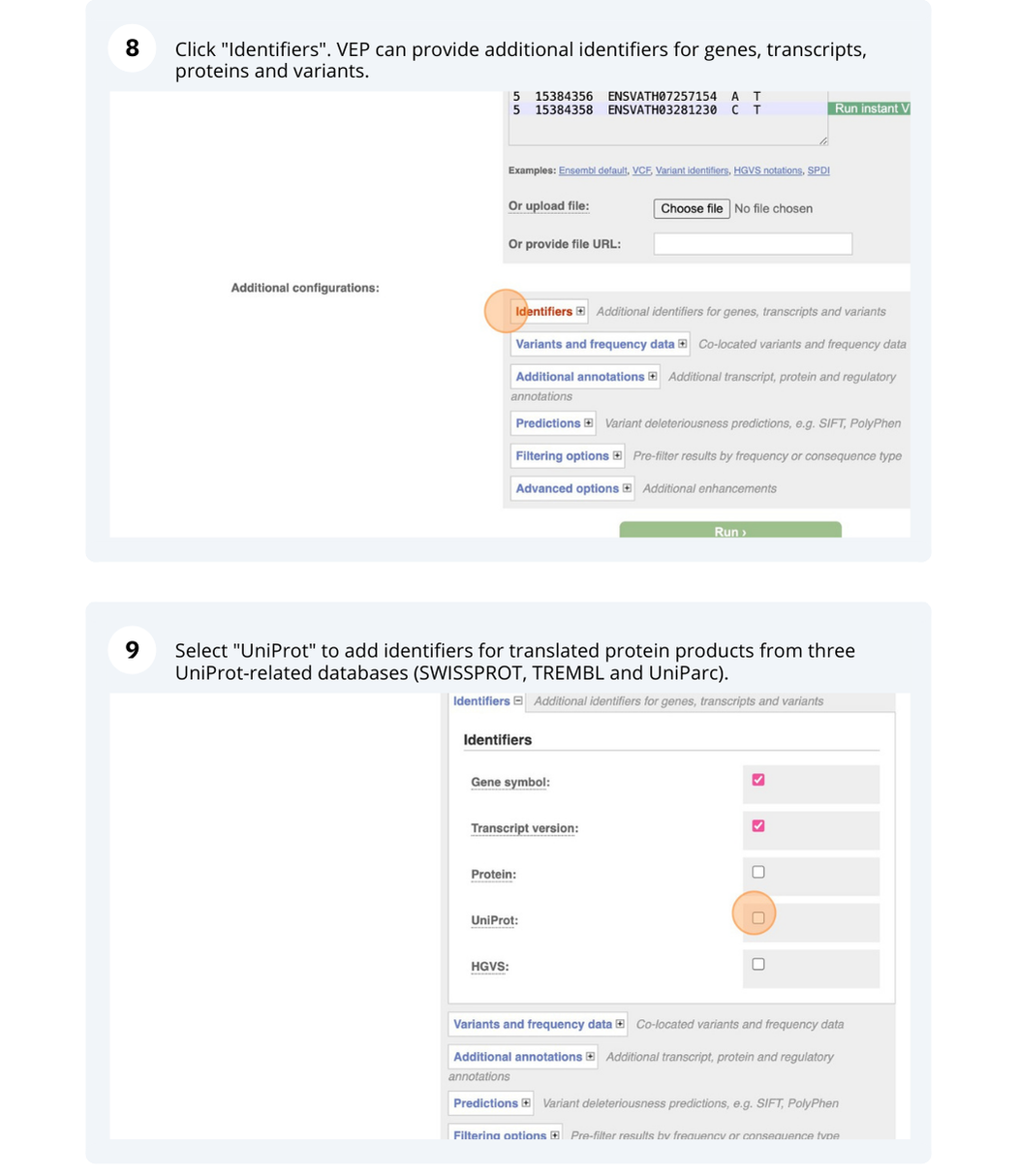
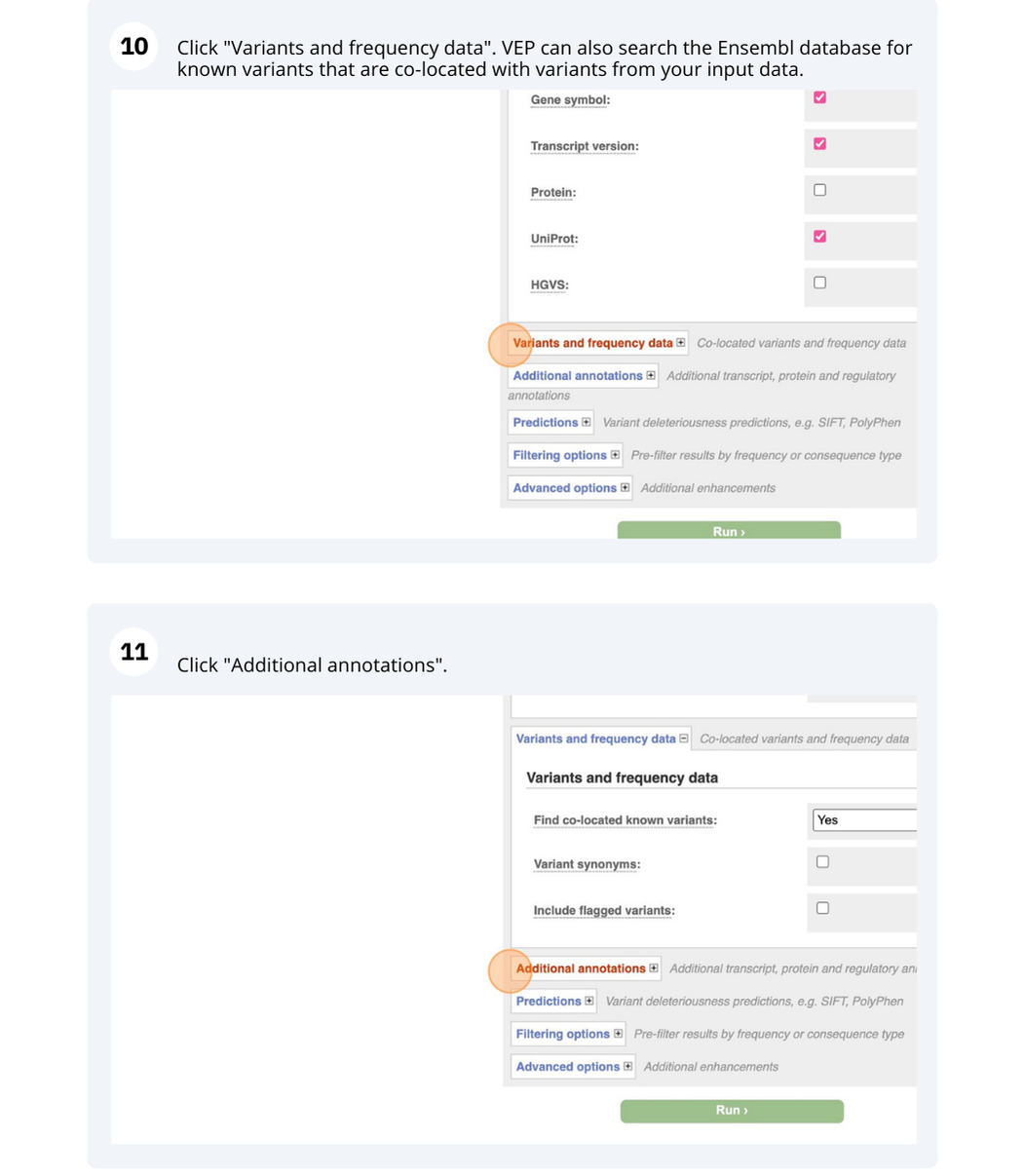
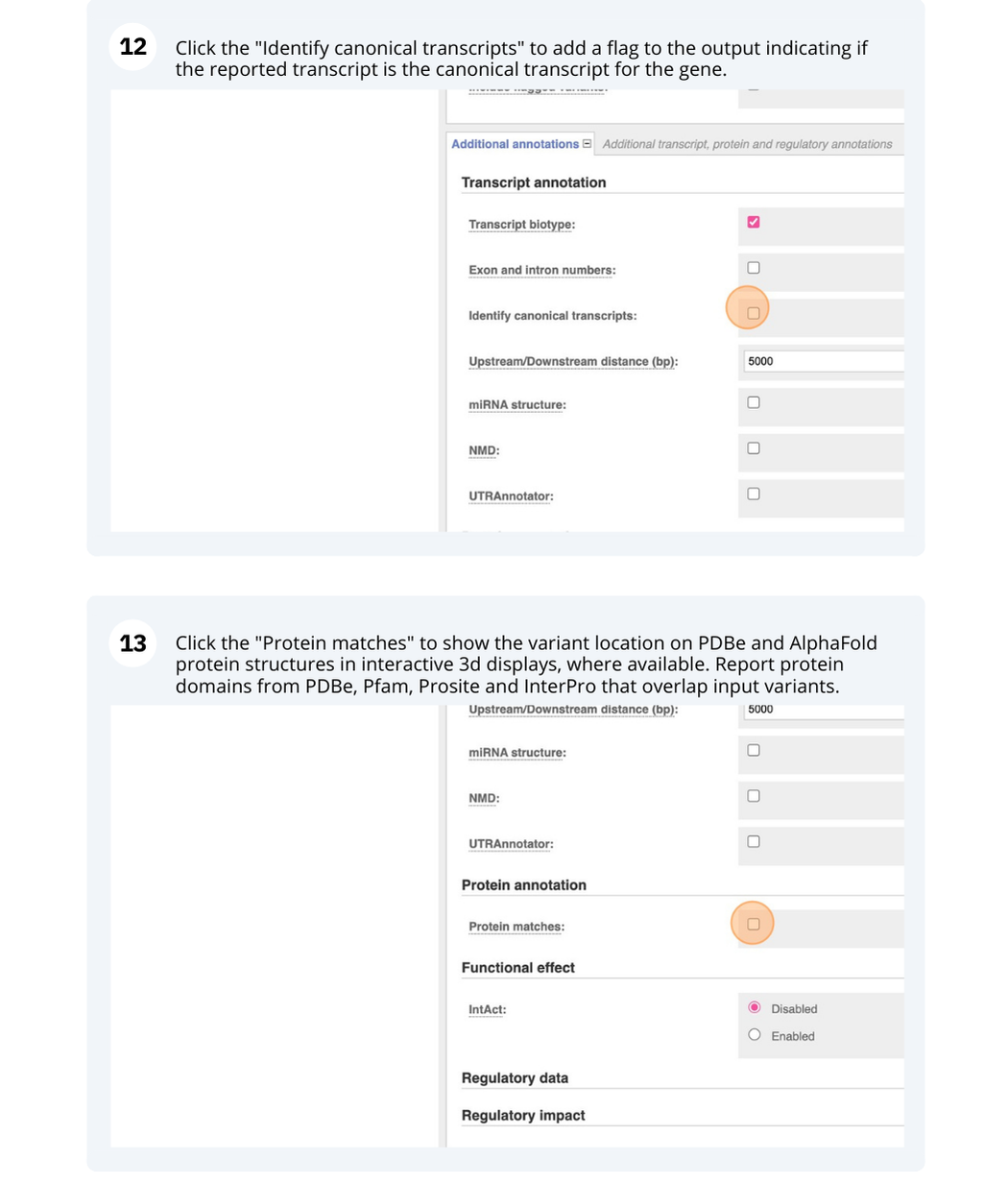
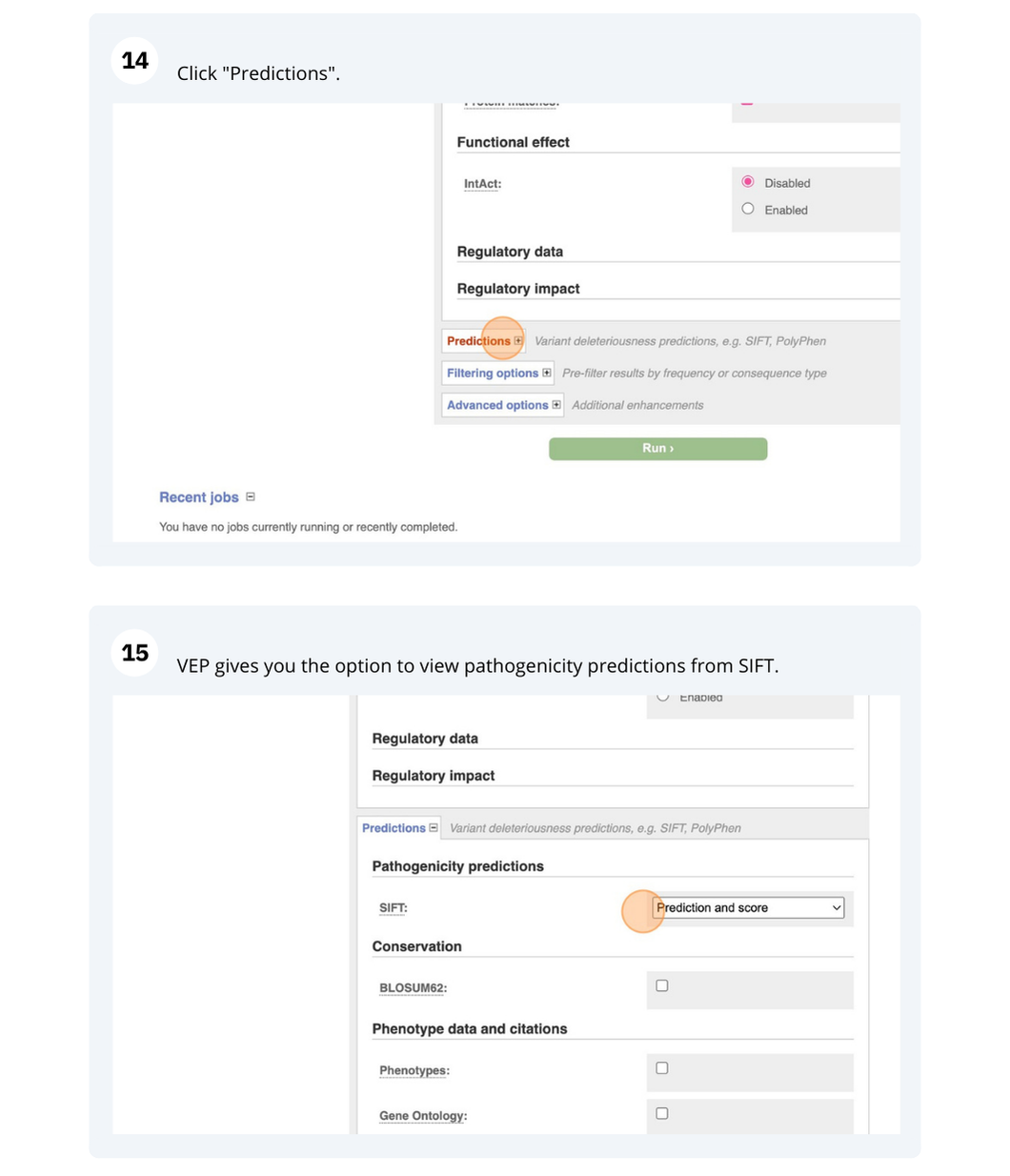
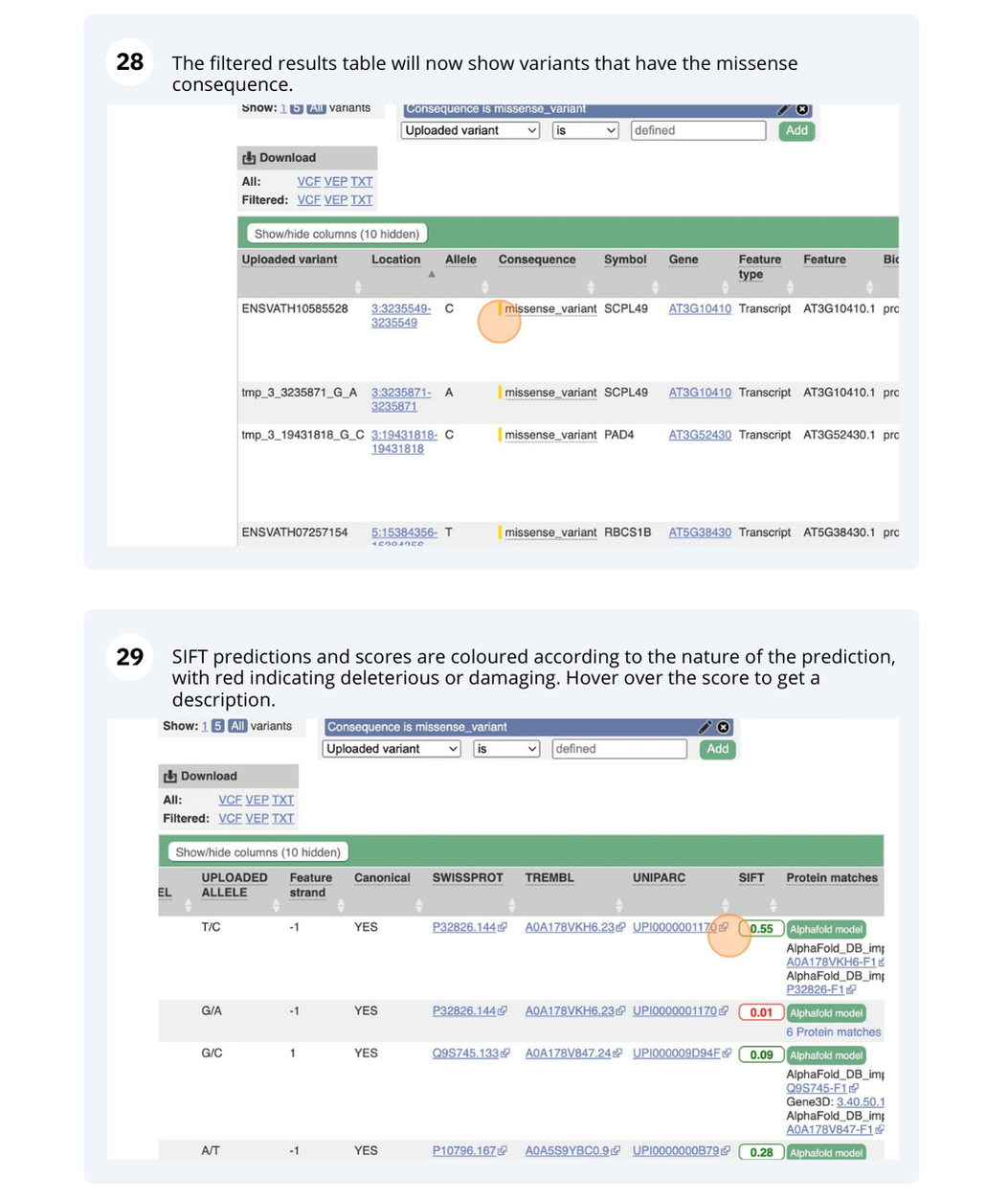
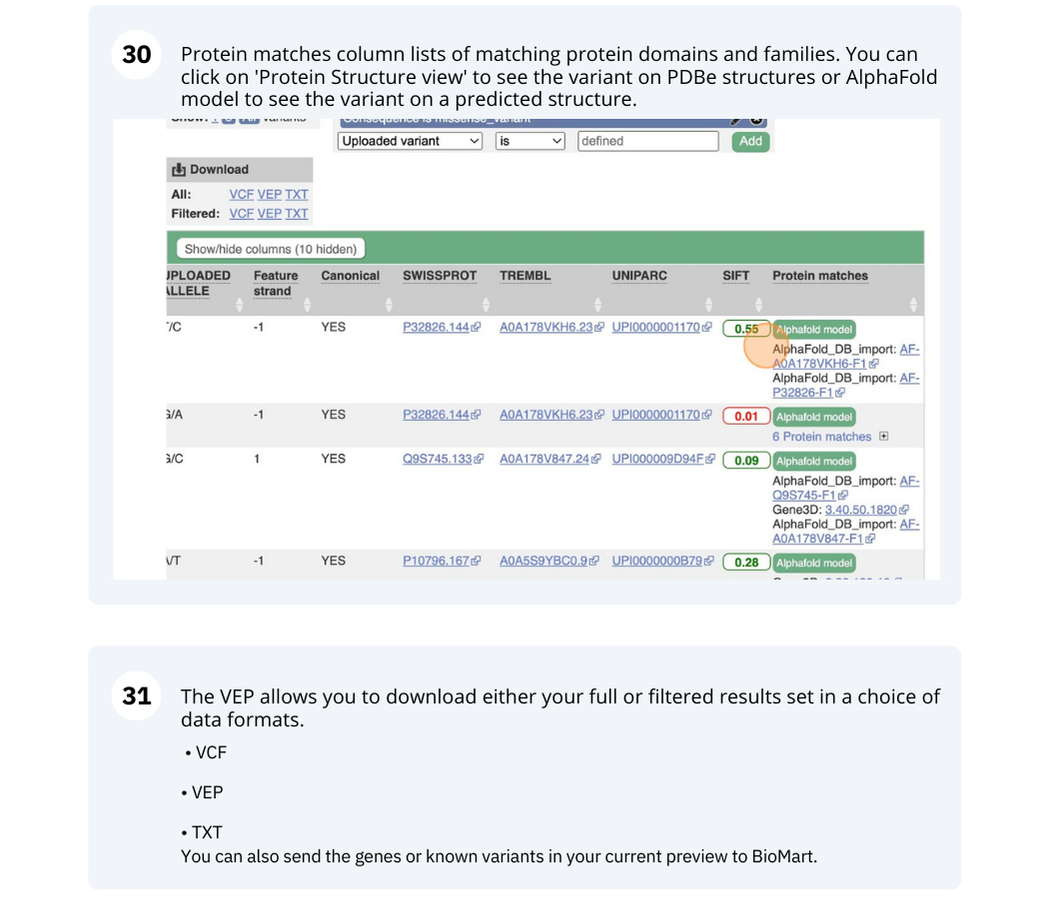
VEP
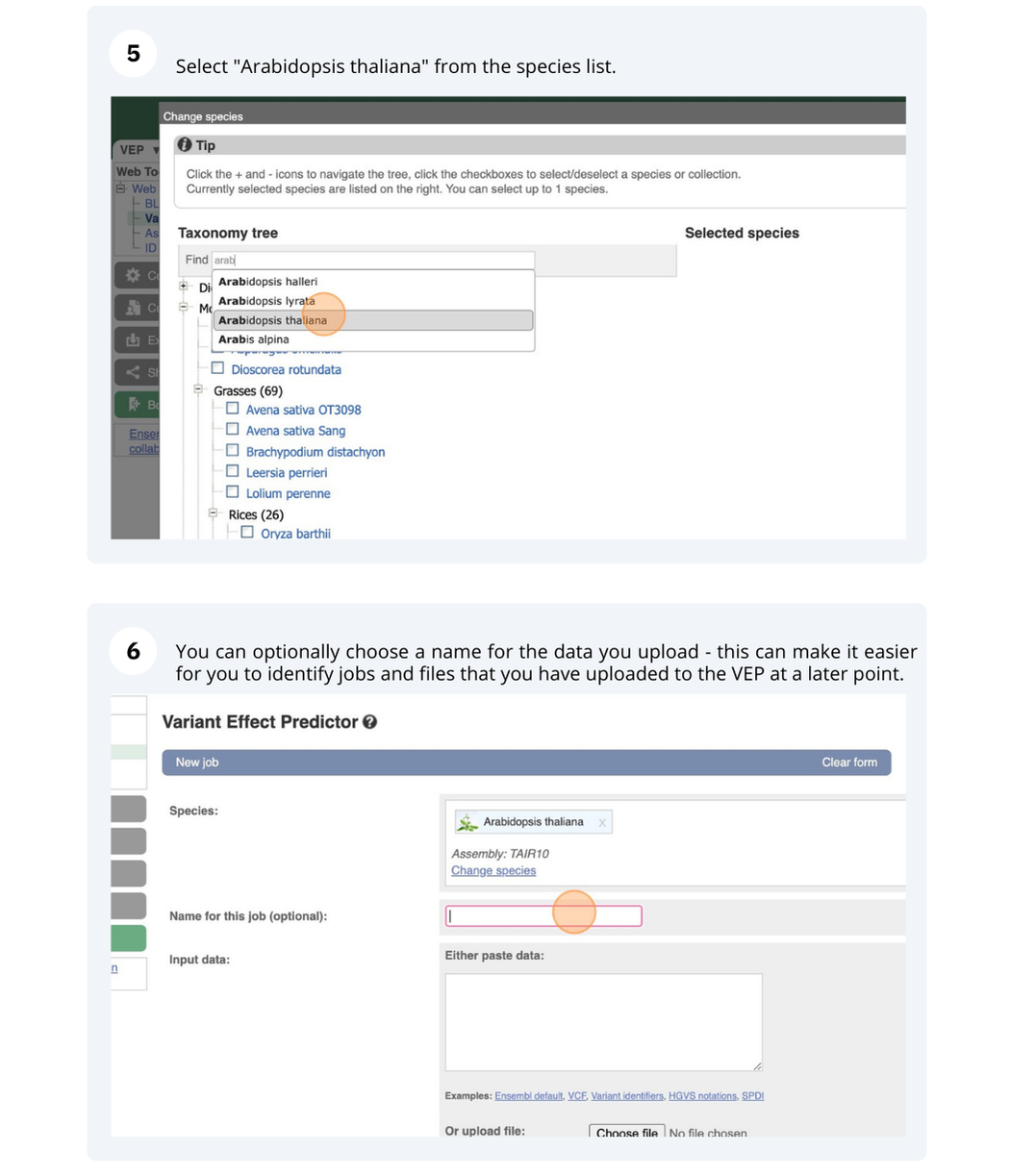
We have identified five variants in Arabidopsis thaliana genome. Use Ensembl VEP to determine if the variants have already been annotated, what genes are affected by these variants and can we get the source and ID for any overlapping protein domains.
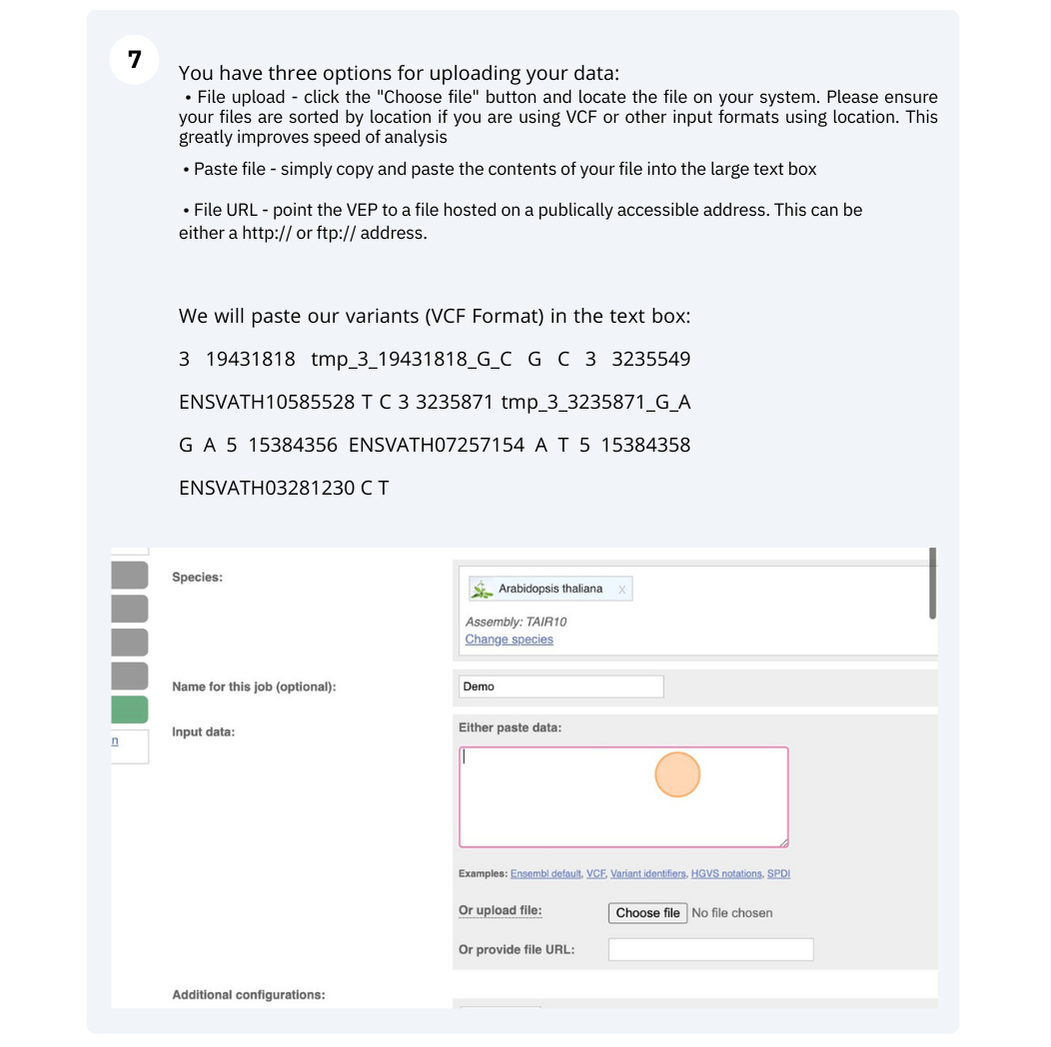
The data is in the VCF format:
Put the following into the Paste data box:
3 19431818 tmp_3_19431818_G_C G C
3 3235549 ENSVATH10585528 T C
3 3235871 tmp_3_3235871_G_A G A
5 15384356 ENSVATH07257154 A T
5 15384358 ENSVATH03281230 C T
VEP analysis of variants in Verticillium dahliae
Verticillium wilt caused by Verticillium dahliae is a notorious soil-borne fungal disease that threatens the yield of economic crops worldwide. We have identified four variants in Verticillium dahliae JR2 chromosome 5:
- C->G at 698711
- G->T at 698935
- G->A at 700313
- C->A at 701484
Use VEP in Ensembl Fungi to answer the following questions:
-
Have these variants already been annotated in Ensembl?
-
What genes are affected by the variants? What are their gene IDs?
-
Are any of the variants predicted to be missense variants?
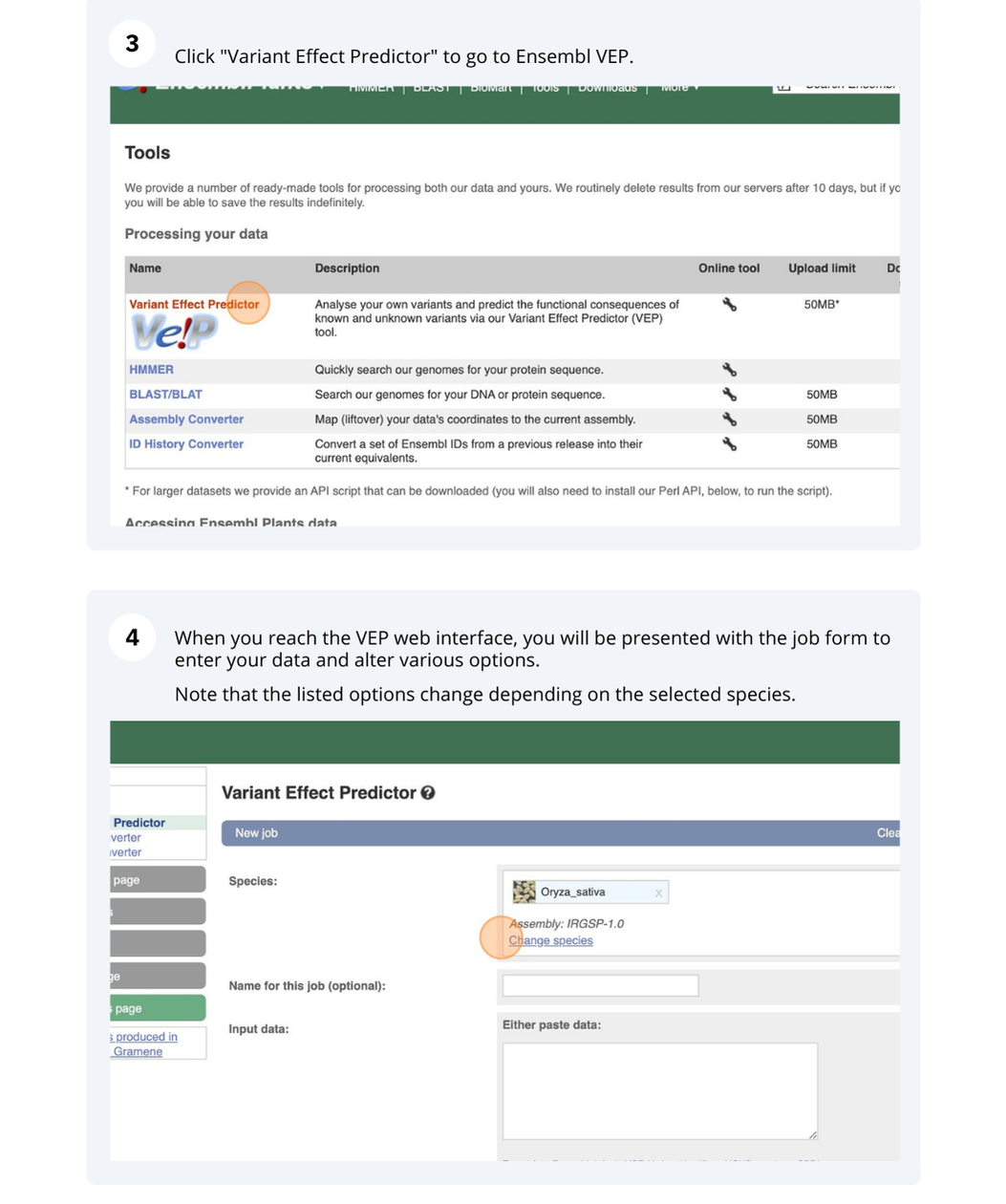
Go to any Ensembl Fungi page and click on Tools in the navigation bar at the top of the page. Click on Variant Effect Predictor and change your species to Verticillium dahliae JR2 by clicking on Change species.
Enter a descriptive name for your VEP job. You will need to convert your variants into one of VEP’s supported input formats. We have converted the variants into the Ensembl default format below. Paste the variants into Input data:.
5 698711 698711 C/G
5 698935 698935 G/T
5 700313 700313 G/A
5 701484 701484 C/A
Click Run at the bottom of the page. When your job is done, click View reesults.
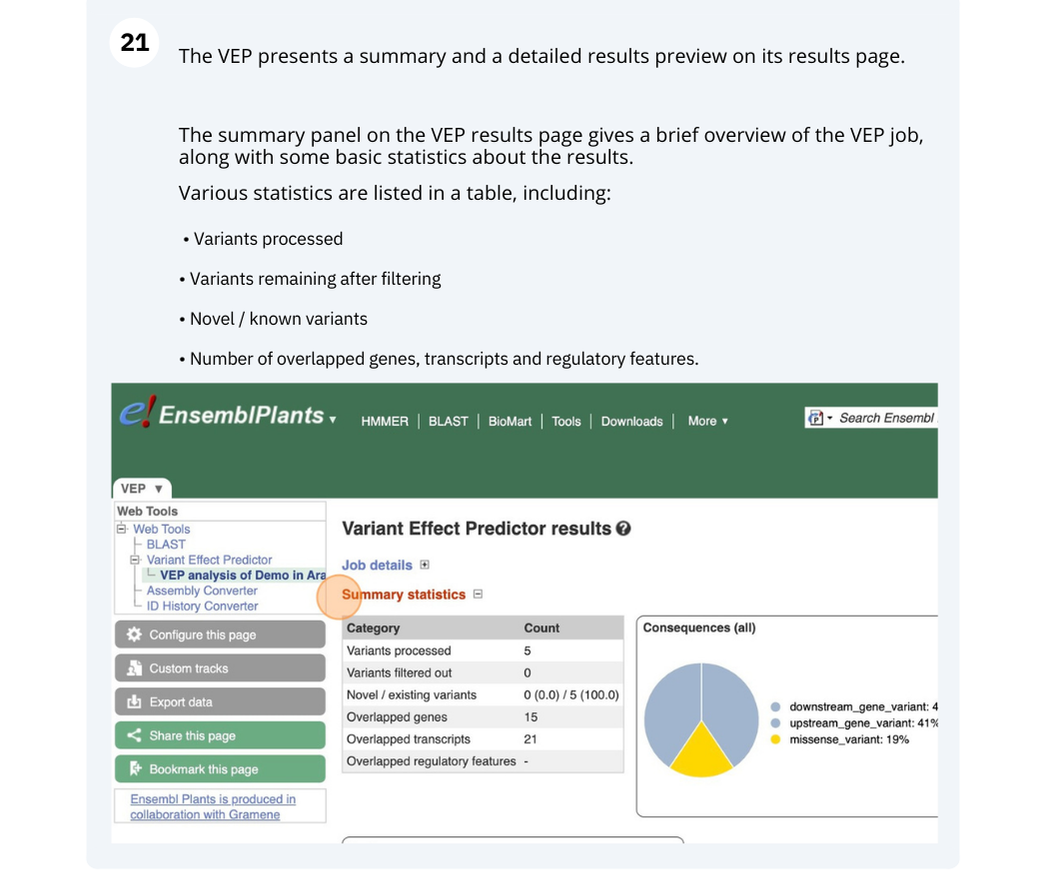
- You can find the number of existing and novel variants in the Summary statistics of the results.
4 variants were analysed, of which 3 are novel.
- You can also find the number of overlapped genes in the Summary statistics.
4 genes are affected.
Sort the table by Gene by clicking on the column name. Count the number of unique gene IDs.
The gene IDs are: VDAG_JR2_Chr5g02150a, VDAG_JR2_Chr5g02160a, VDAG_JR2_Chr5g02170a and VDAG_JR2_Chr5g02171a.
- Filter the table as follows: Consequence
is missense_variant.Yes, the third variant (5_700313_G/A) is predicted to have a missense effect on gene VDAG_JR2_Chr5g02170a.
Web VEP analysis of variants in Oryza sativa Japonica (rice)
You’ll find a VCF file here. This is a small subset of the outcome of Oryza sativa Japonica whole-genome sequencing and variant-calling experiment. Analyse the variants in this file with the VEP tool in Ensembl Plants and determine the following:
-
How many genes and transcripts are affected by variants in this file?
-
Do these variants result in a change in the proteins encoded by any of the Ensembl genes? Which genes are affected? What is the amino acid change? What is the pathogenicity prediction score for this change?
Go to Ensembl Plants and click on Tools at the top of the page. Click on Variant Effect Predictor and select Oryza sativa Japonica Group from the Species menu.
Either click on Choose file and select the file to upload it, or directly paste the URL into the Or provide file URL: box. Click Run at the bottom of the page. When your job is done, click View results.
- The number of affected genes and transcripts is shown in the Summary statistics table at the top.
8 genes and 8 transcripts are affected by these variants.
- Use the filters to view only missense variants. The filters are found above the detailed results table in the middle. Select Consequence and is from the drop-down menus. Then type
missense_variantinto the boxe. Add to apply your filter.1 variant is a missense variant. It causes a leucine to arginine (L/R) at position 16 change in the gene OS09G0103500. The SIFT score is 0.01 (Deleterious low confidence). Refere to this link for more information on SIFT (https://sift.bii.a-star.edu.sg/).
Comparative Genomics
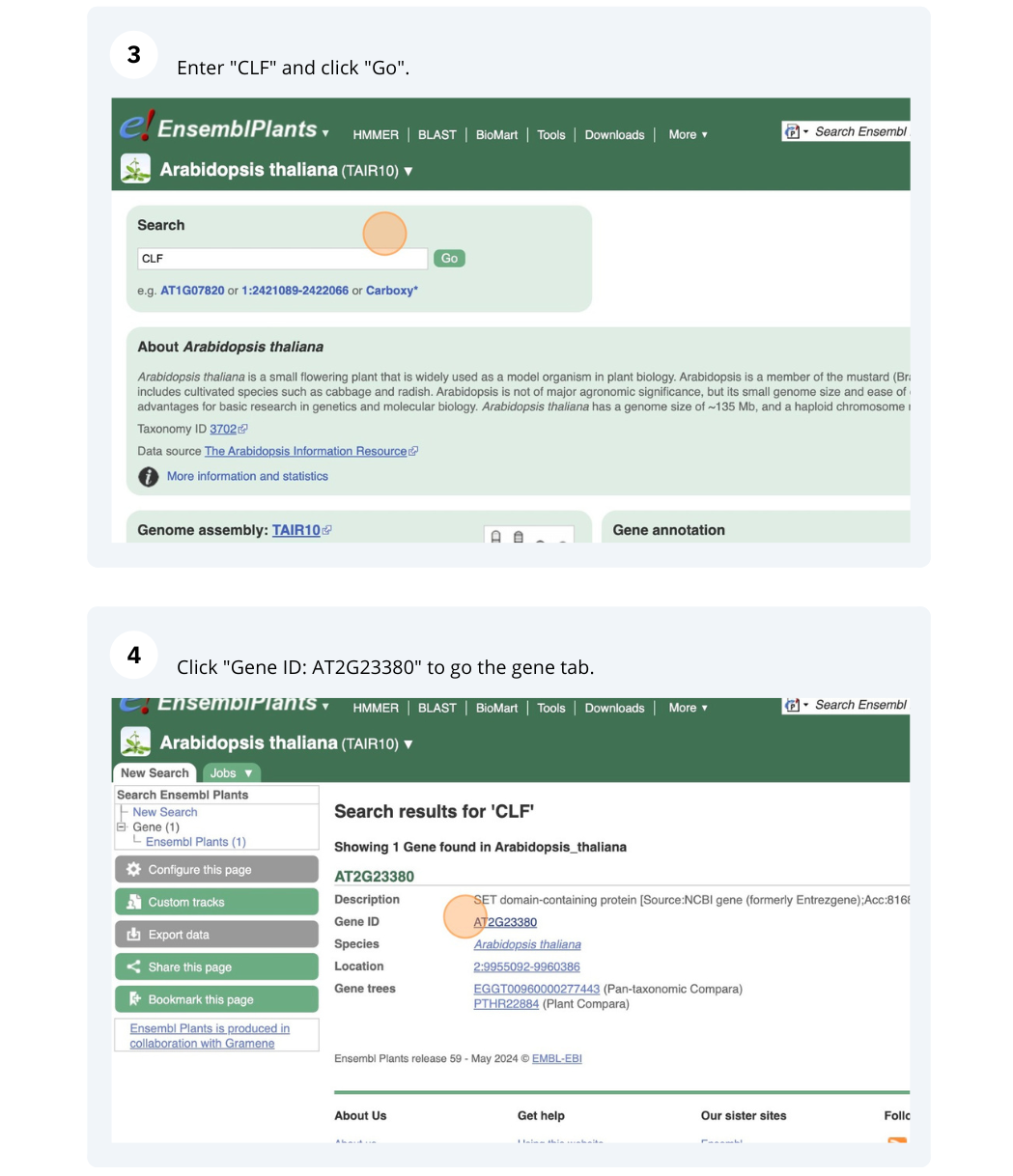
The CURLY LEAF (CLF) gene of the model plant Arabidopsis thaliana is involved in the control of leaf and flower morphology and flowering time. We will use Ensembl Plants to look at the homologues of Arabidopsis thaliana gene CLF (AT2G23380) and genomic alignment with other species.
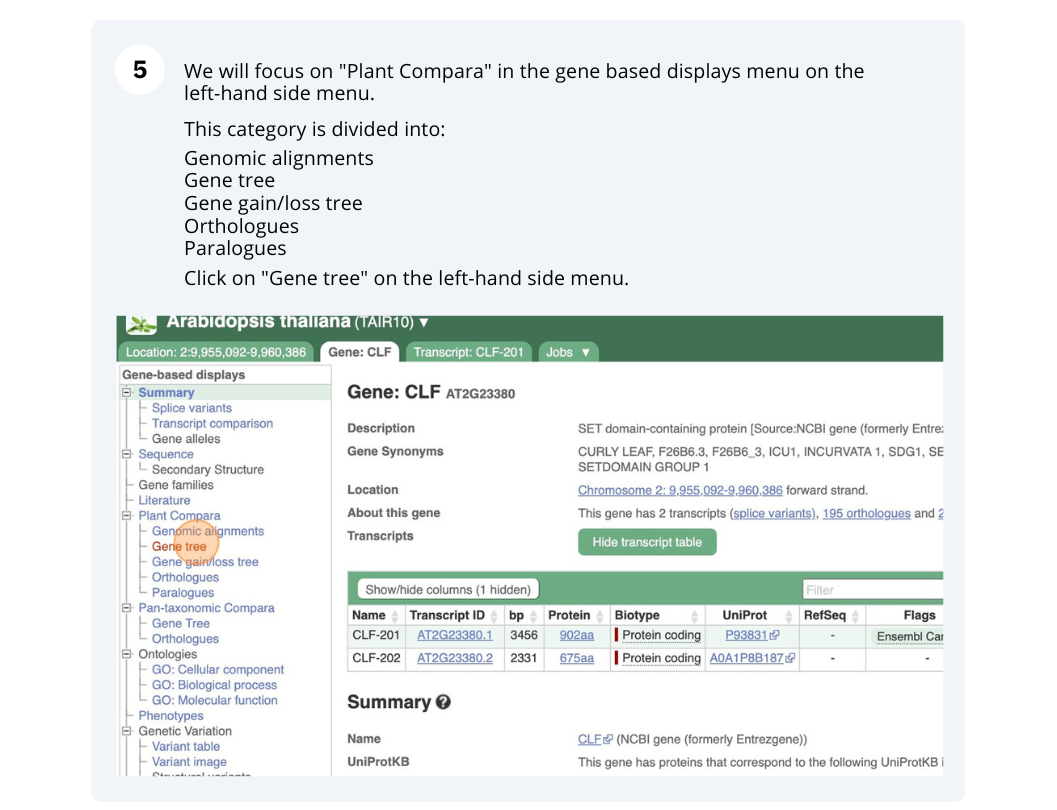
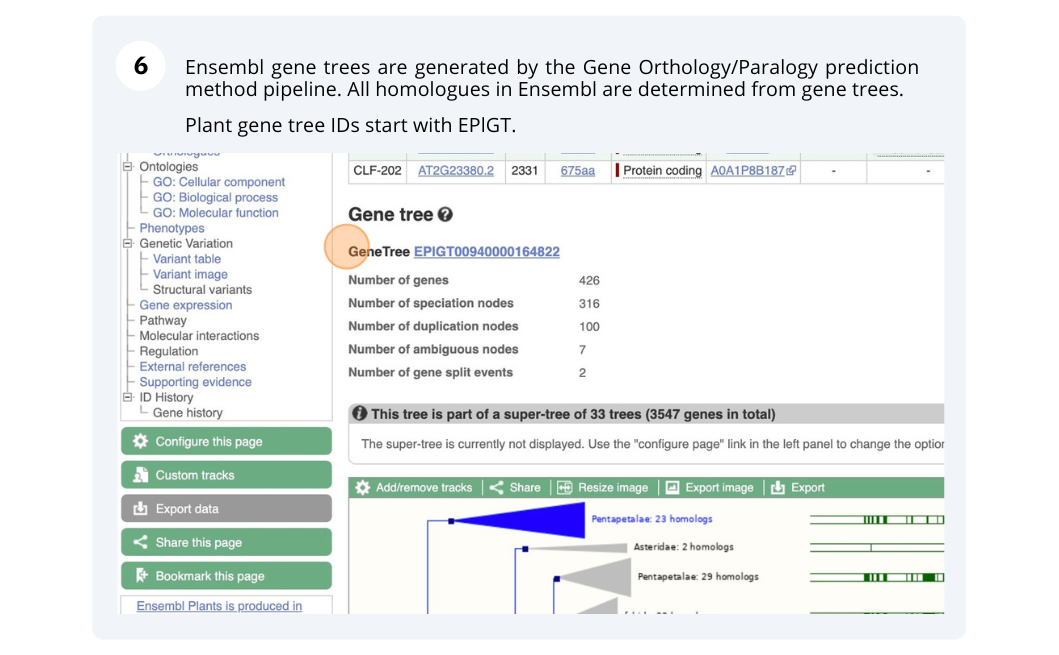
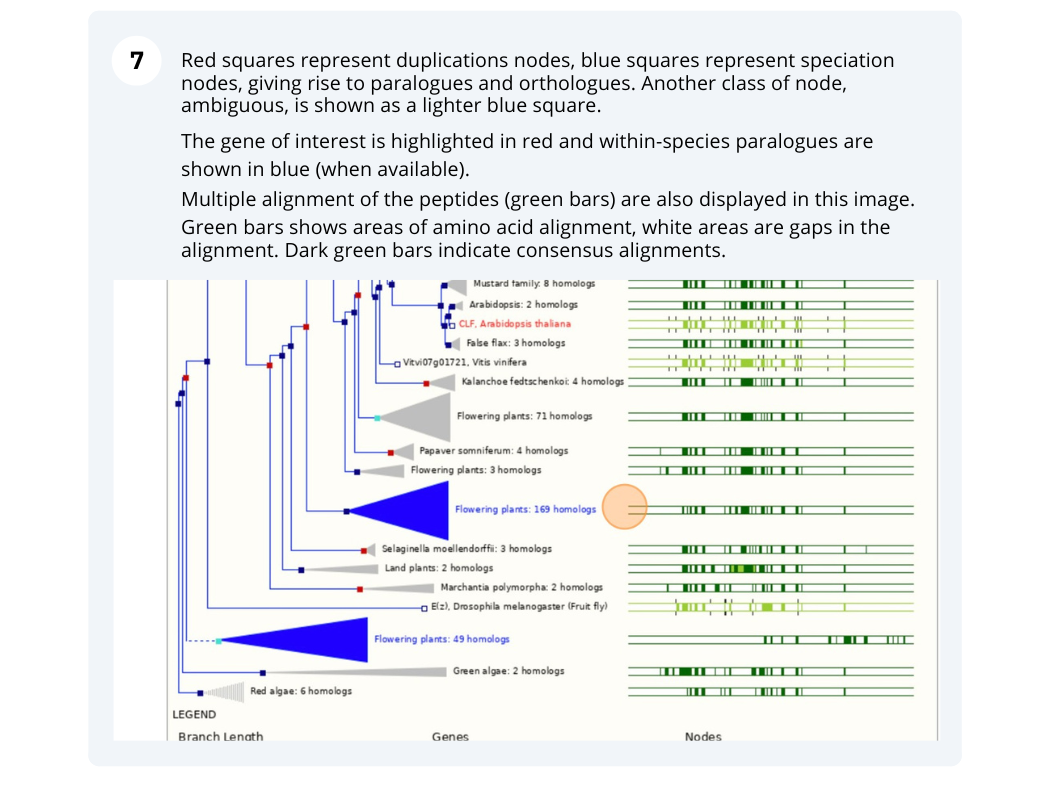
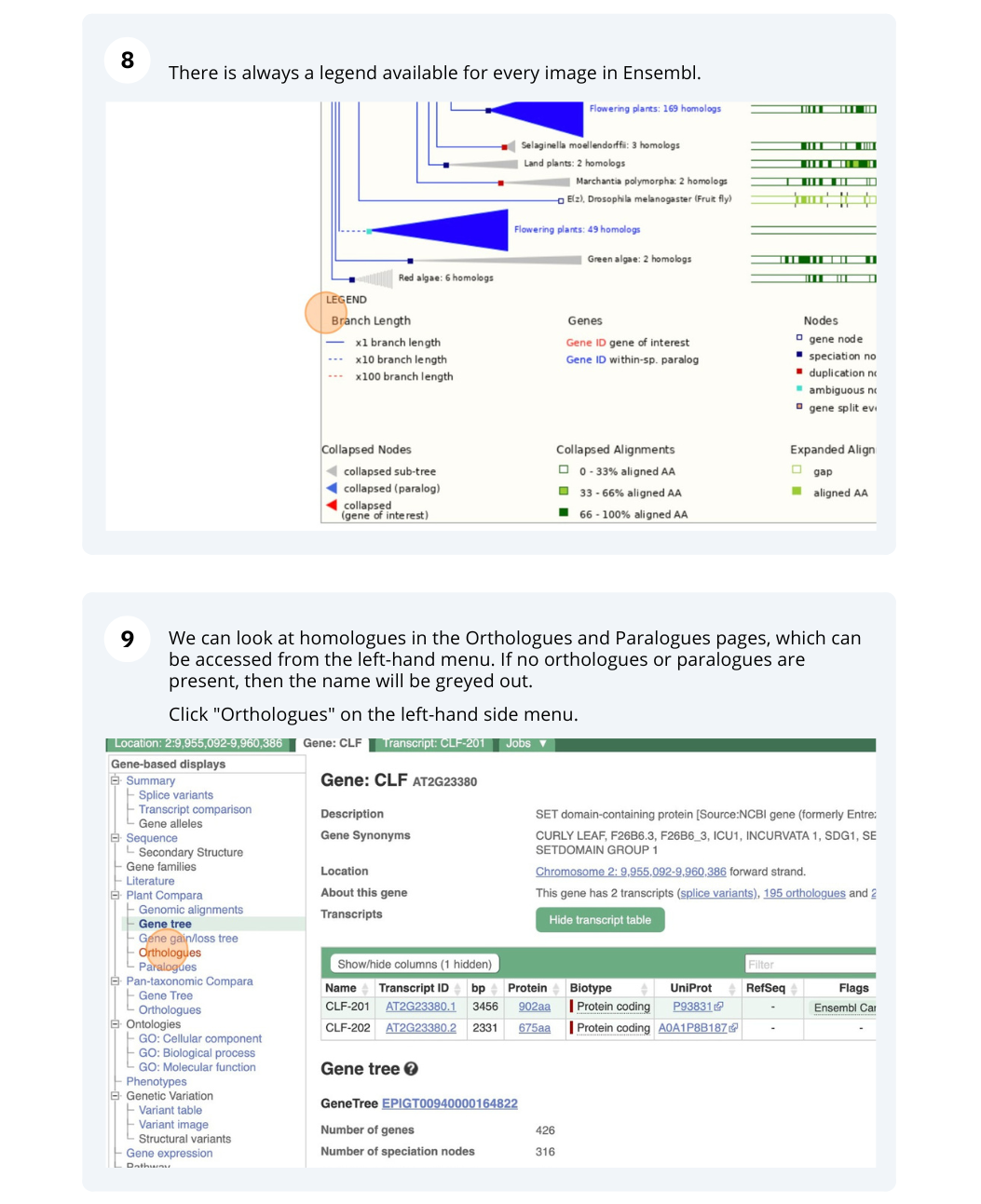
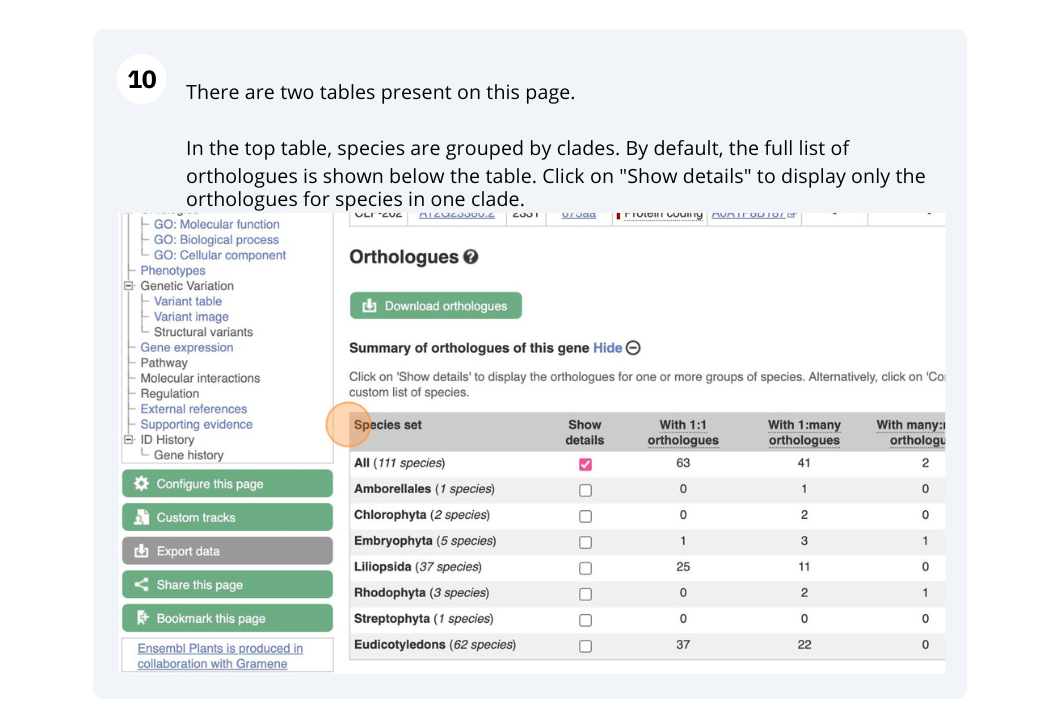
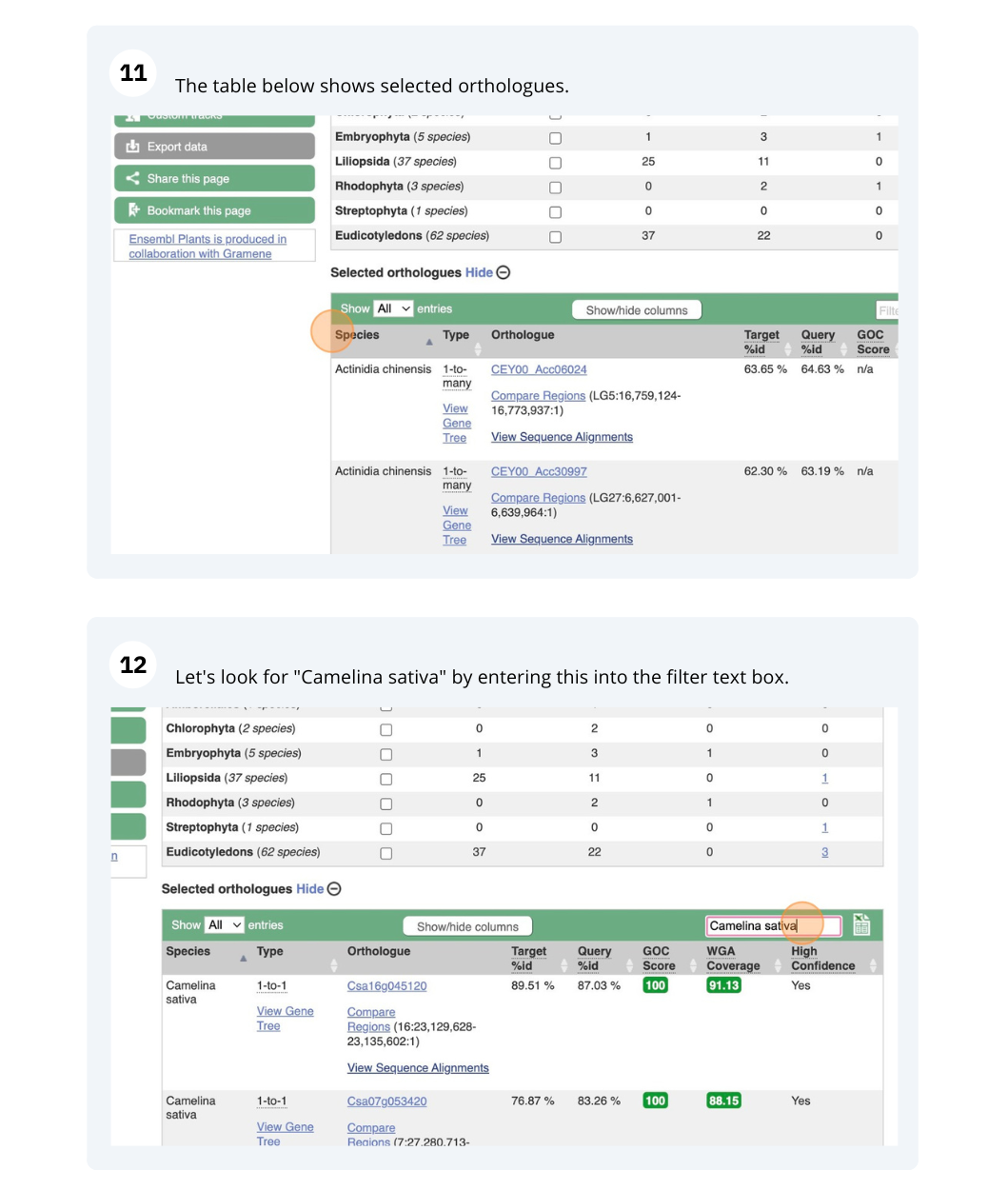
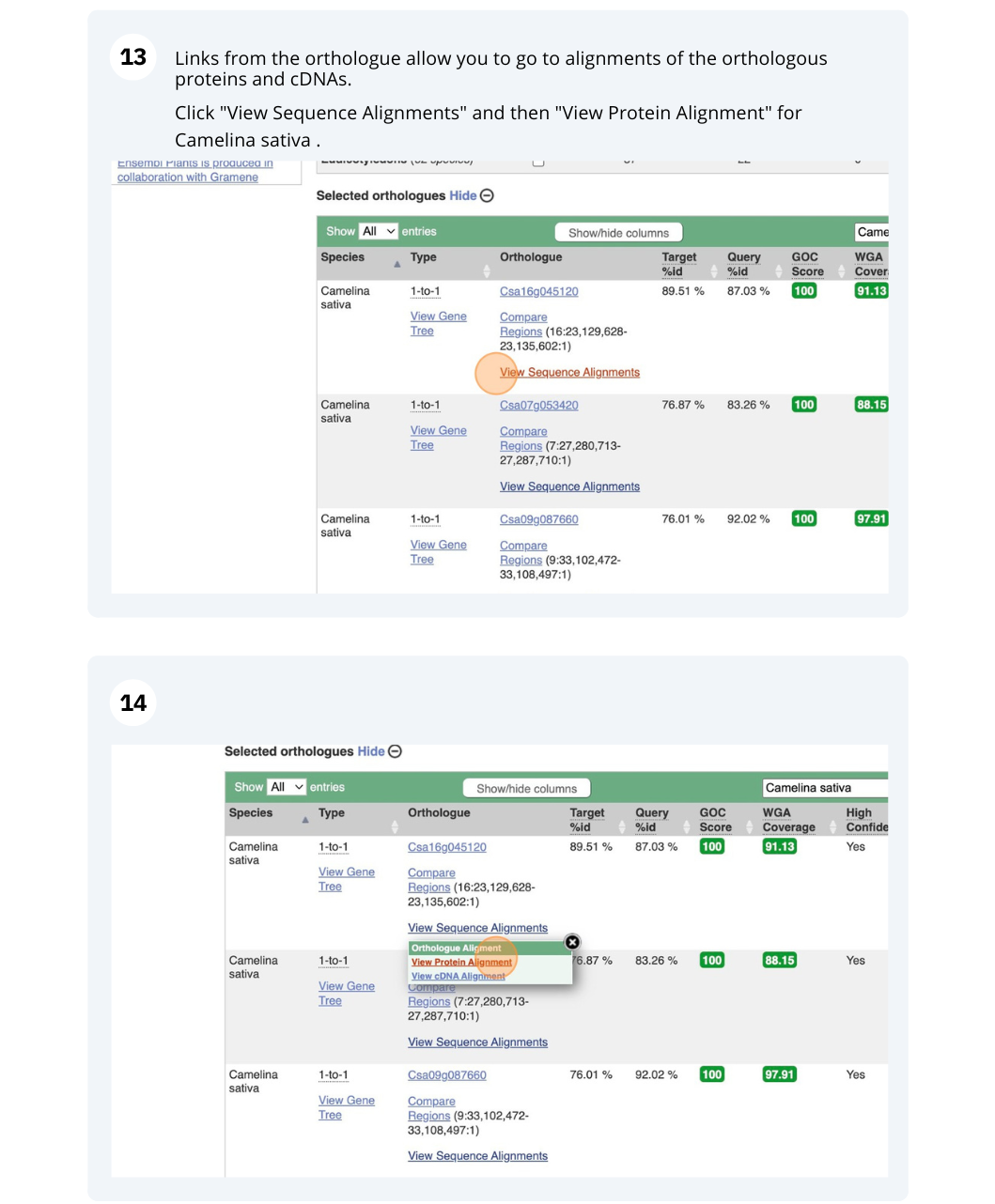
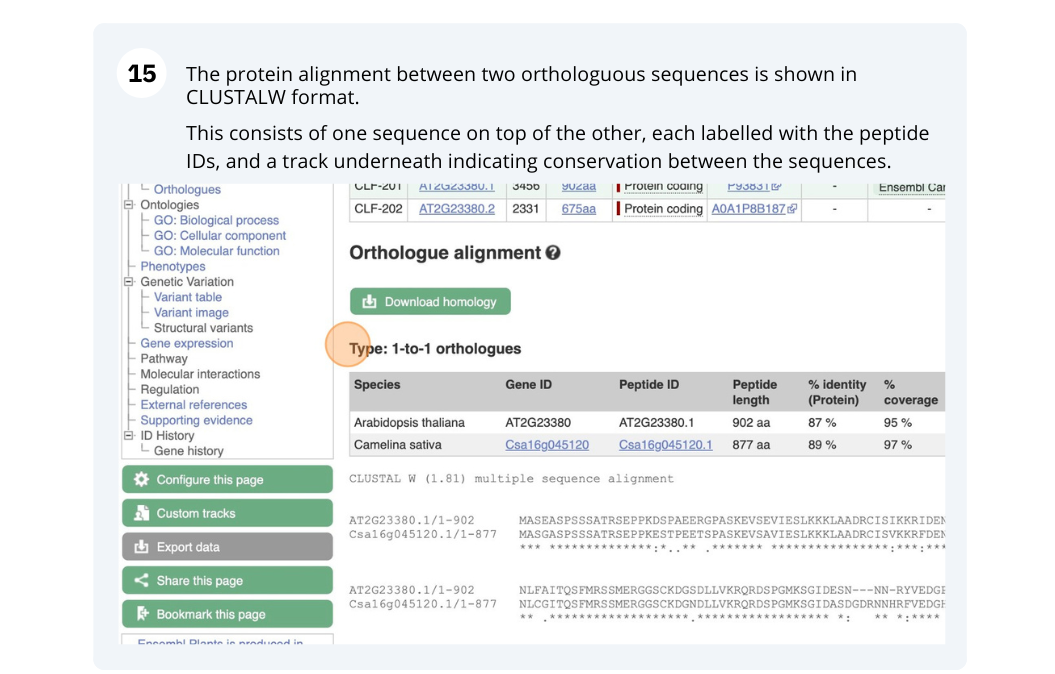
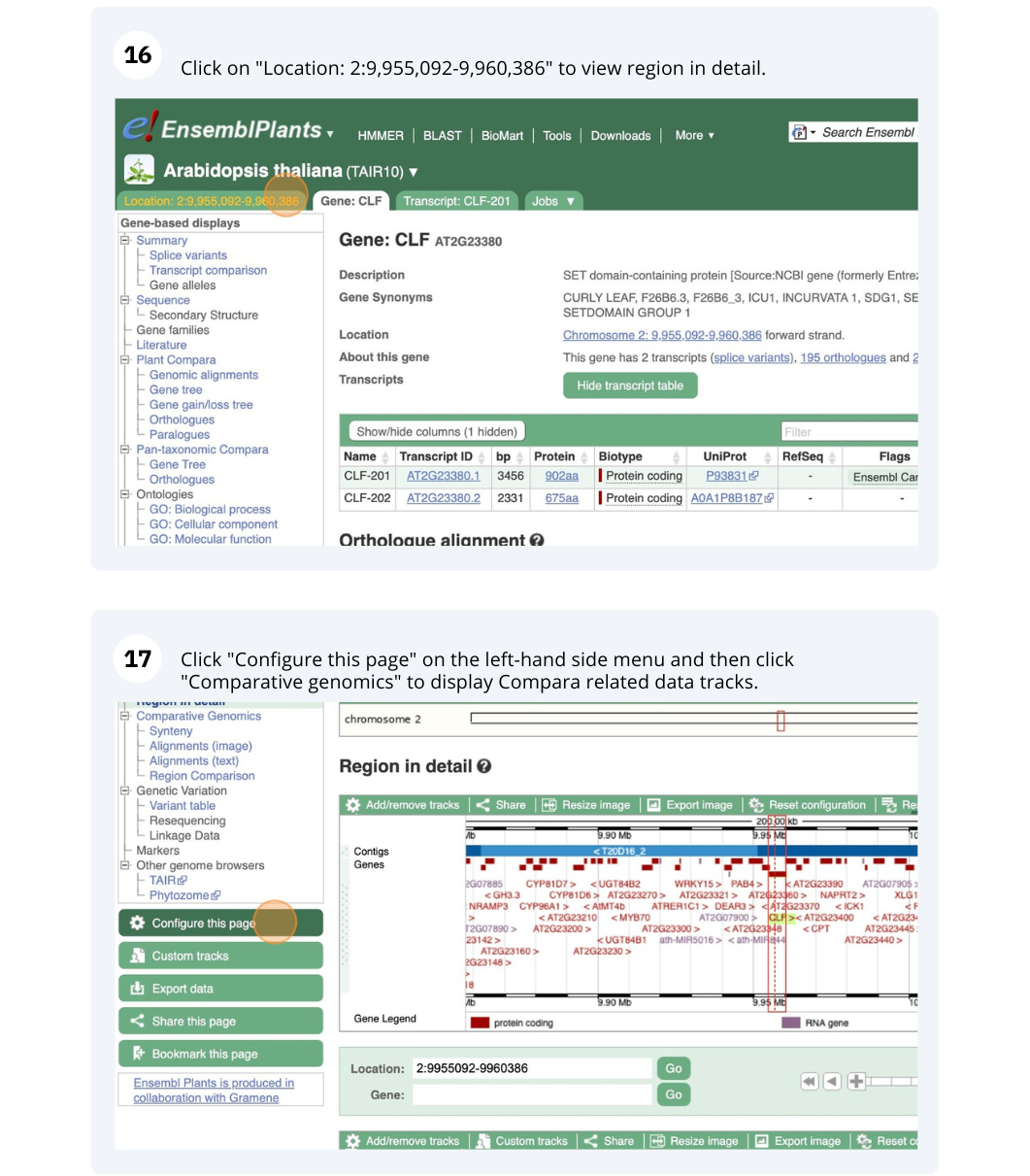
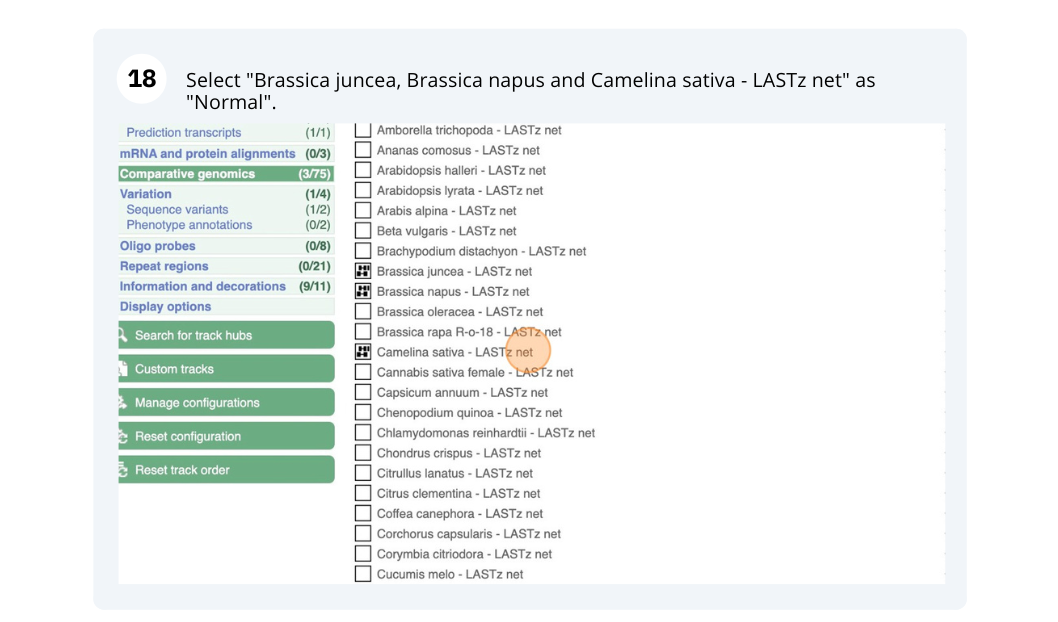
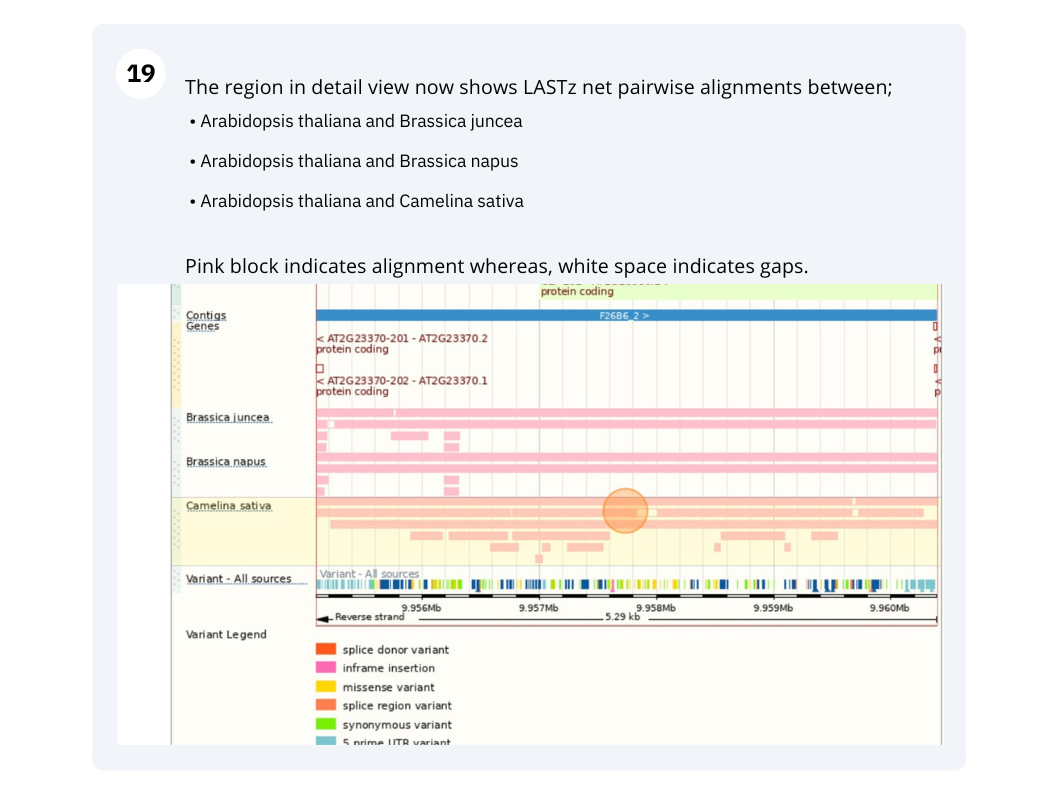
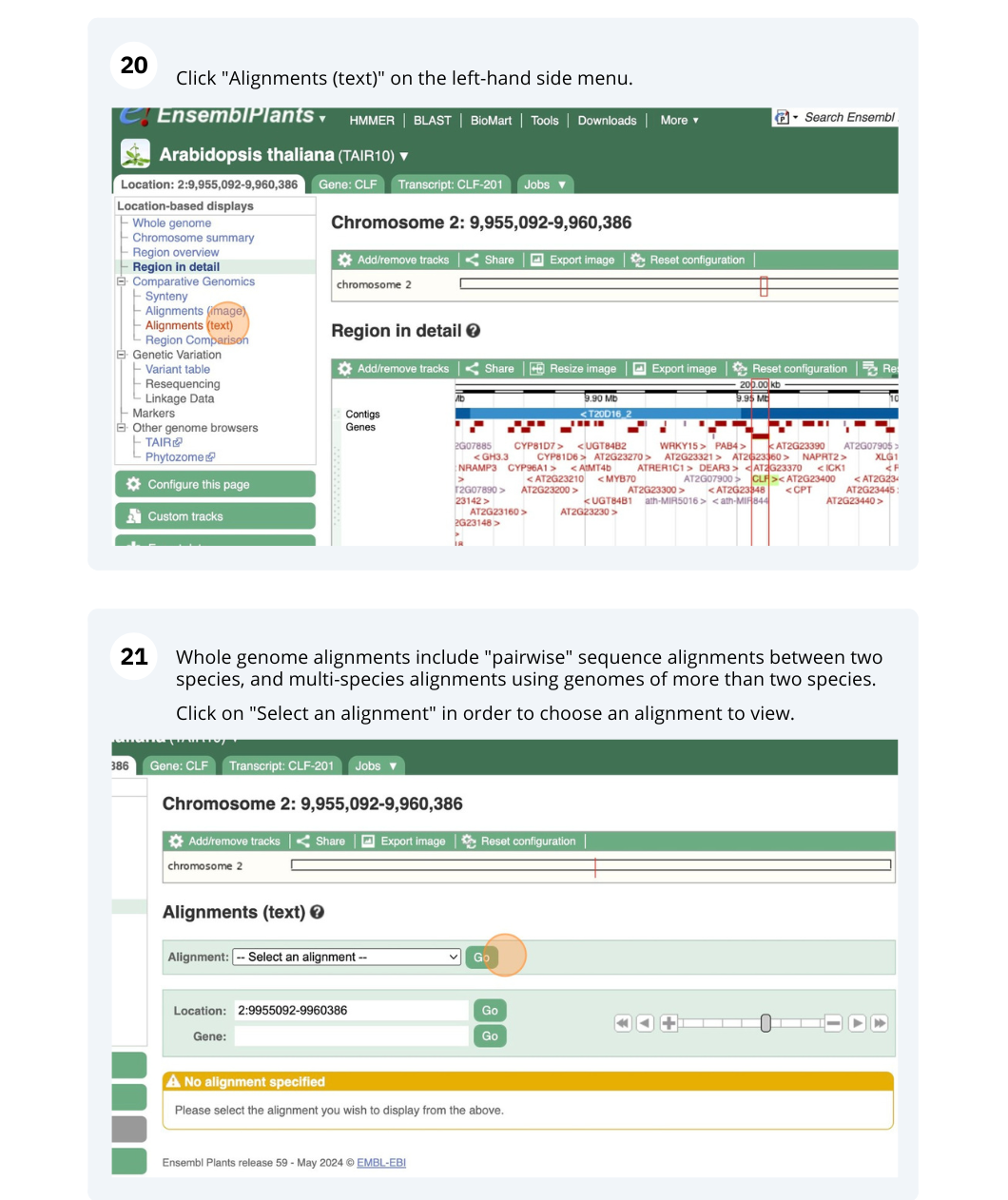
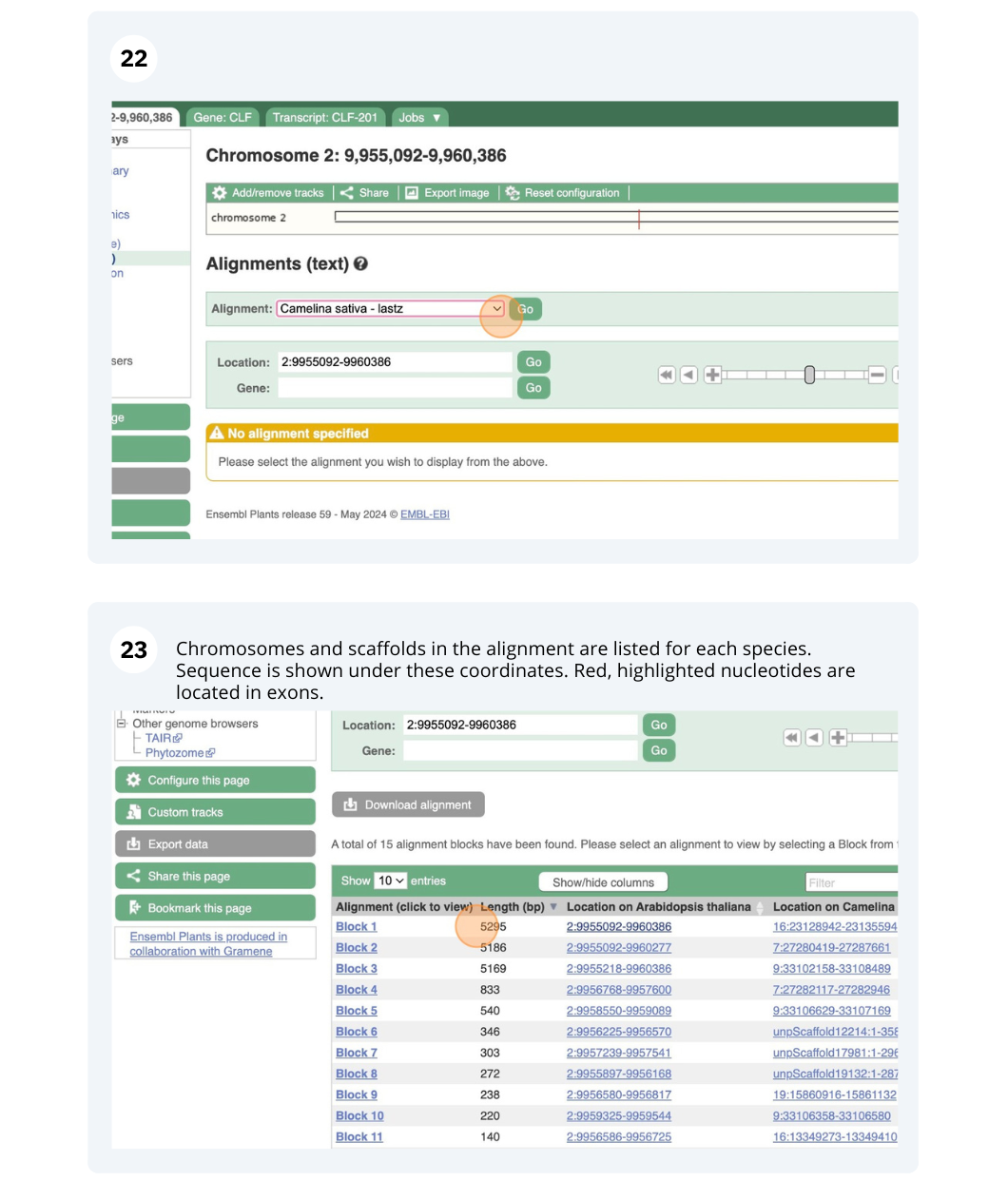
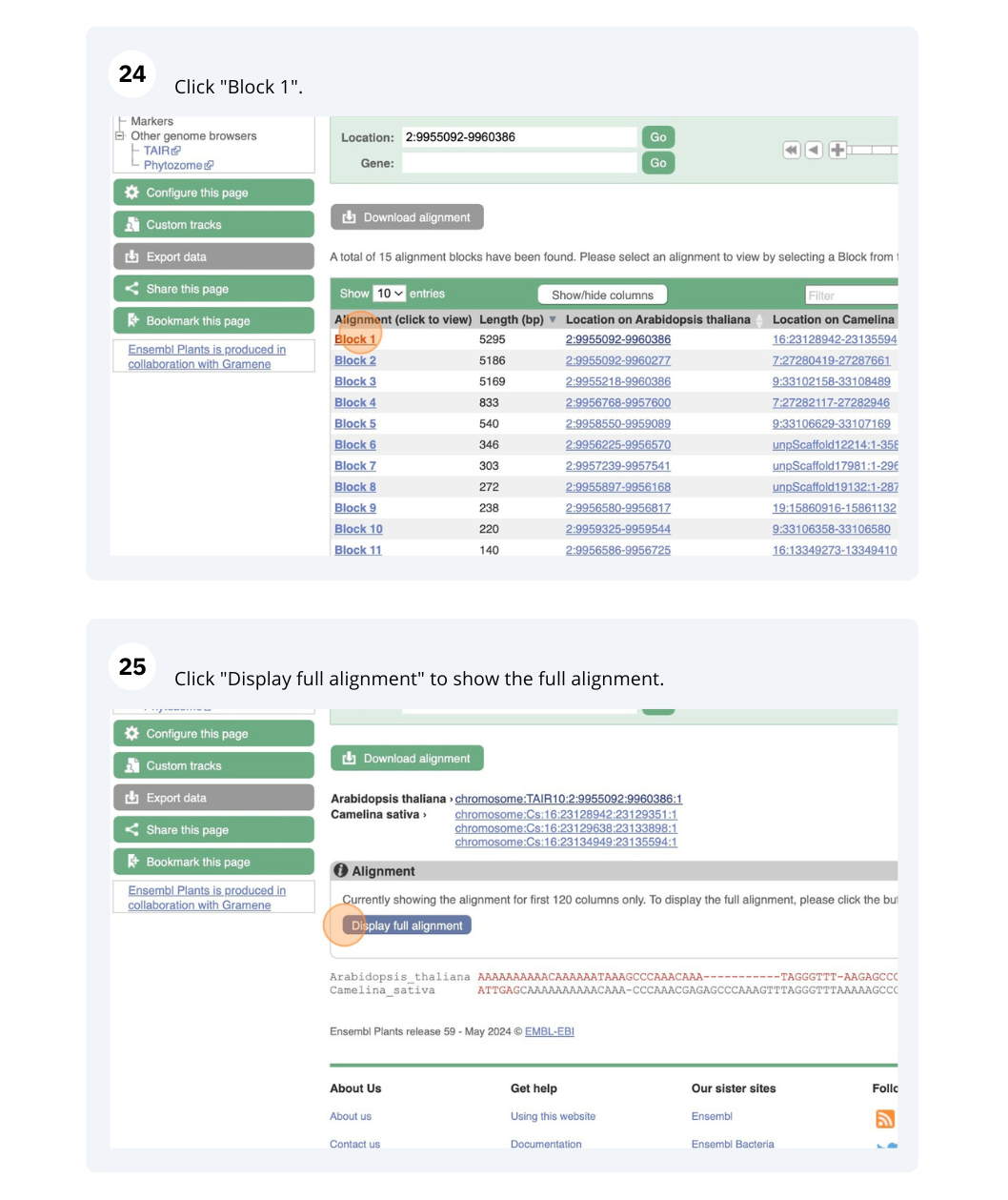
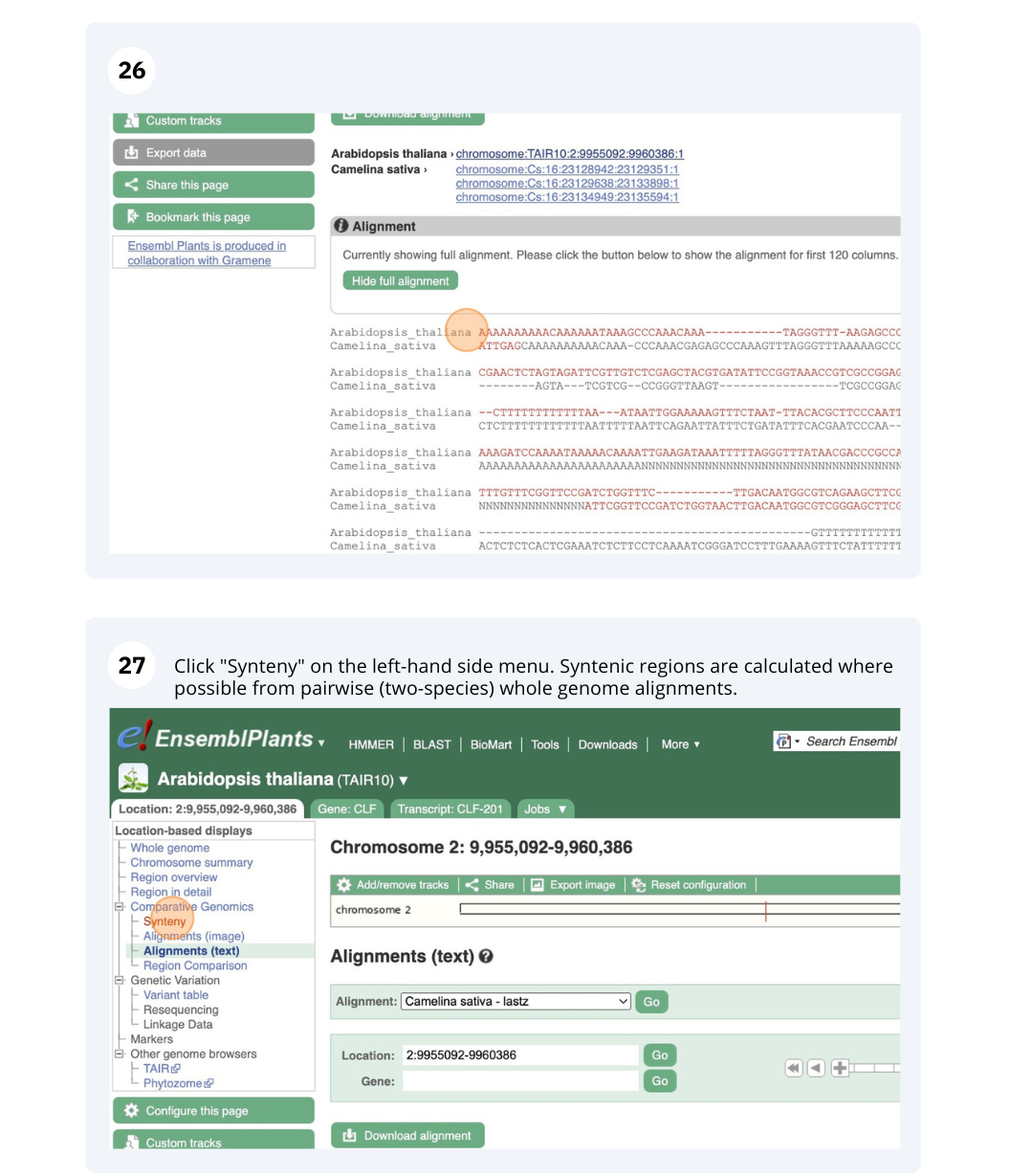
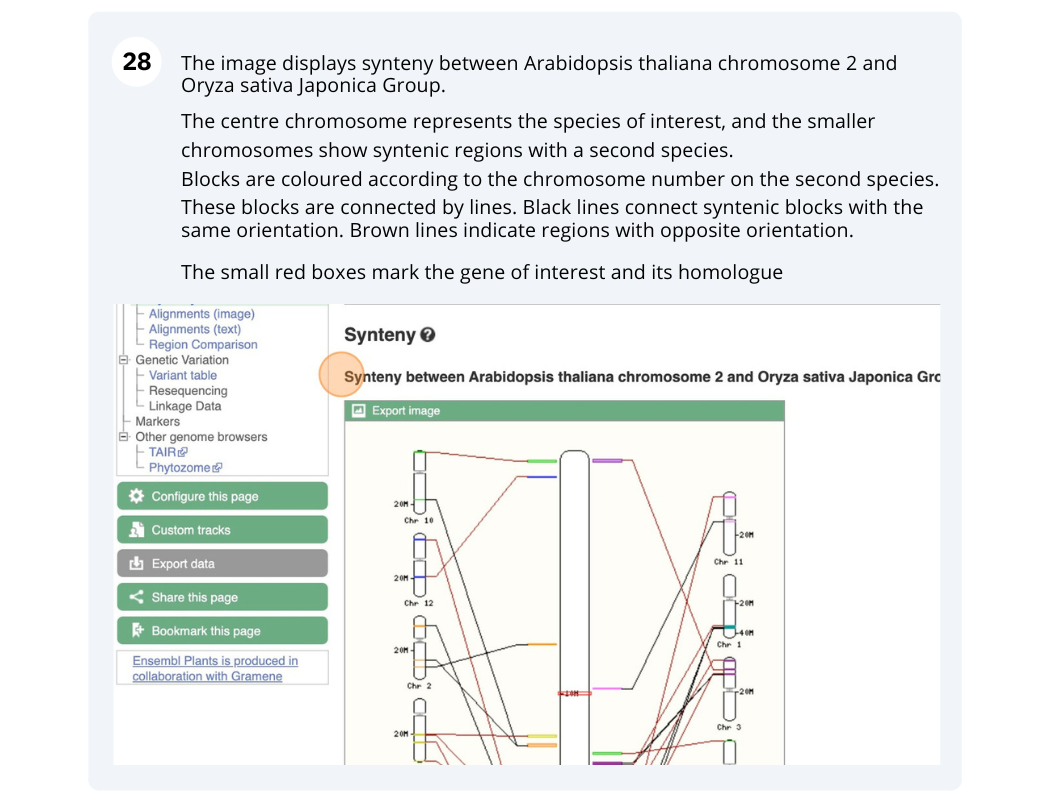
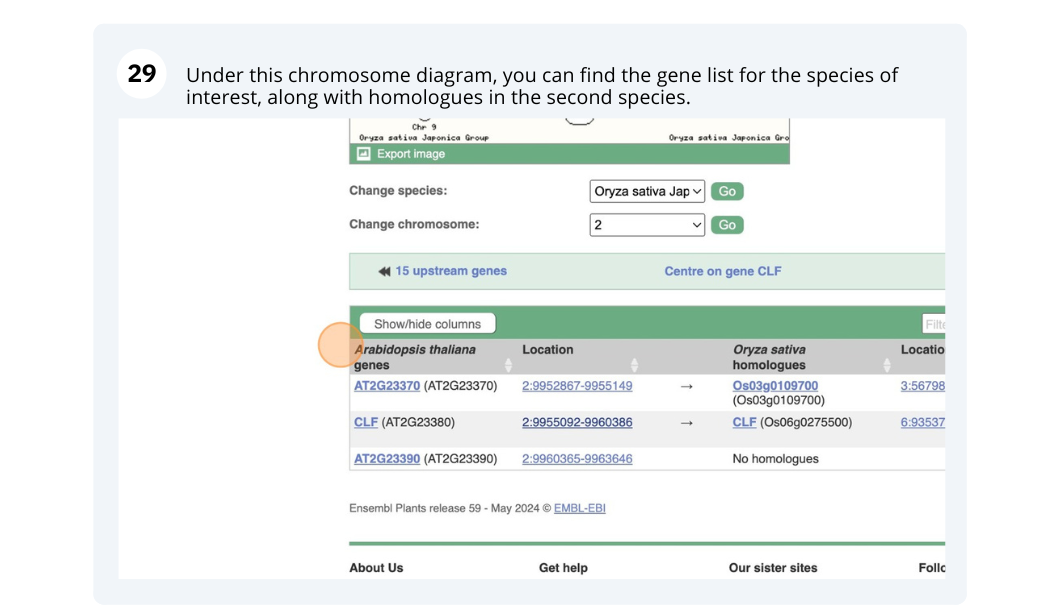
Finding orthologous genes for a root transporter in Oryza sativa Japonica (rice)
Search Ensembl Plants for the gene Lsi1 in Oryza sativa Japonica Group (rice). This gene is known to code for an aquaporin transporter that facilitates the uptake of silicon and arsenic through the roots. Silicon concentration is highest in grass species, and is associated with defence.
-
From the gene tab, go to the Orthologues page under Plant Compara. Which plant group has the highest number of 1-to-1 orthologues? Is it the same group that has the highest number of 1-to-many orthologues?
-
Reduce the orthologues table to look only at Triticum aestivum (wheat) orthologues. Why are there three results for a 1-to-1 orthologue?
-
Click on the Compare regions link for chromosome 6B region in wheat to go to the Location tab. Scroll to the bottom image. How do the gene models compare between the species? Do they have the same number of exons?
-
Click back to the Gene tab and click on the Gene gain/loss tree page. Which species has the highest number of members of this gene family? Is it a grass? Can you change the view to see a radial tree?
Go to Ensembl Plants. Look for the main search box highlighted in green. Select Oryza sativa Japonica Group from the drop-down box and type in Lsi1. Click Go and click on the gene ID Os02g0745100.
- Go to Plant Compara: Orthologues on the left-hand panel.
Liliopsida has 24 1-to-1 orthologues, the only group with 1-to-1 orthologues. This group is synonymous with Monocotyledon, so the group that contains the grasses. Eudicotyledons has the highest number of 1-to-many orthologues, indicating that this gene has been duplicated in the eudicots.
- Use the search box in the top right-hand corner of the Selected orthologues table and enter
Triticum aestivum, the table should automatically filter.There are 3 results, one for each component (A,B,D). Note that these are considered 1-to-1 orthologues, rather than 1-to-many. This is because these genes arose in wheat by hybridisation (allopolyploidy), rather than duplication (autopolyploidy).
- Click on Compare regions (found in the 3rd column below the gene identifier) from the 2nd result for component 6B. This takes us to the Location tab. Scroll down to the bottom of the page.
Both genes have 5 exons and the same structure. This looks unusual because the gene in rice is on the forward strand, while the gene in wheat is on the reverse strand. This is reflected in the crossing green links between the pink alignment blocks.
- Click on the Gene tab at the top of the page and click on Gene gain/loss tree in the left-hand panel.
Significant expansions are shown with red branches, and the number of genes in the family shown in the count next to the image and species name. We can see that Echinochloa crus-galli (Cockspur grass) has 25 members in this group.
We can change the tree to radial view by clicking on the icon with two arrows at the top left of the image.
BioMart
The BioMart tool can be used to export customized datasets directly from the Ensembl and Ensembl Genomes databases. It allows the extraction of the same data for a list of annotated features (genes, transcripts, variants, genomic regions).
Follow these instructions to guide you through BioMart to answer the following query:
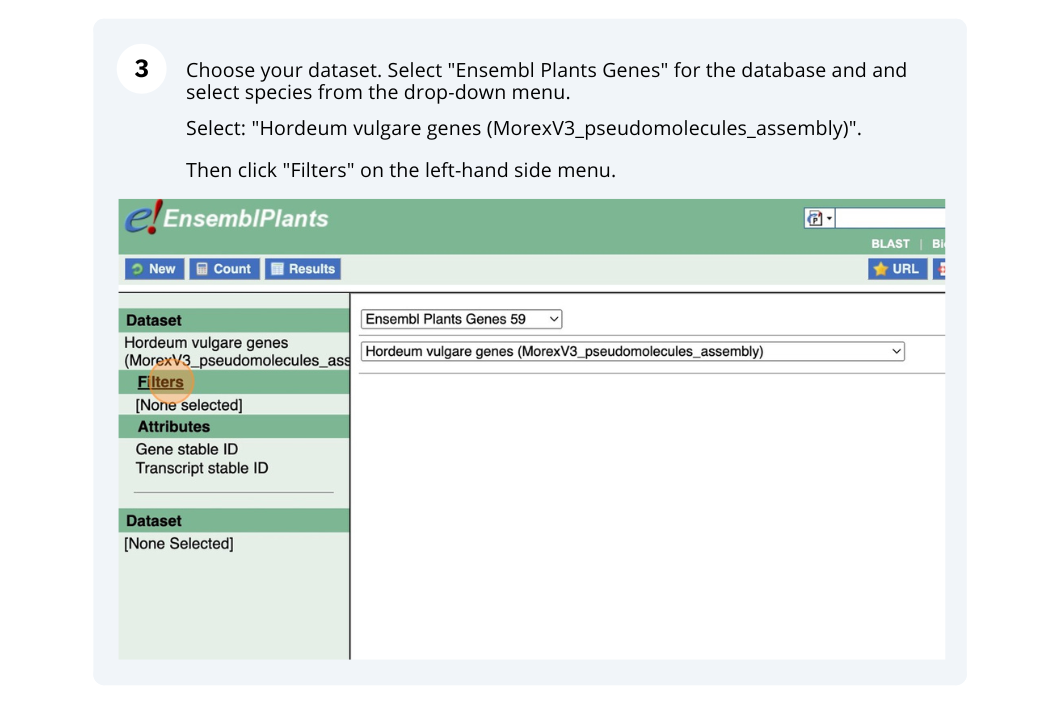
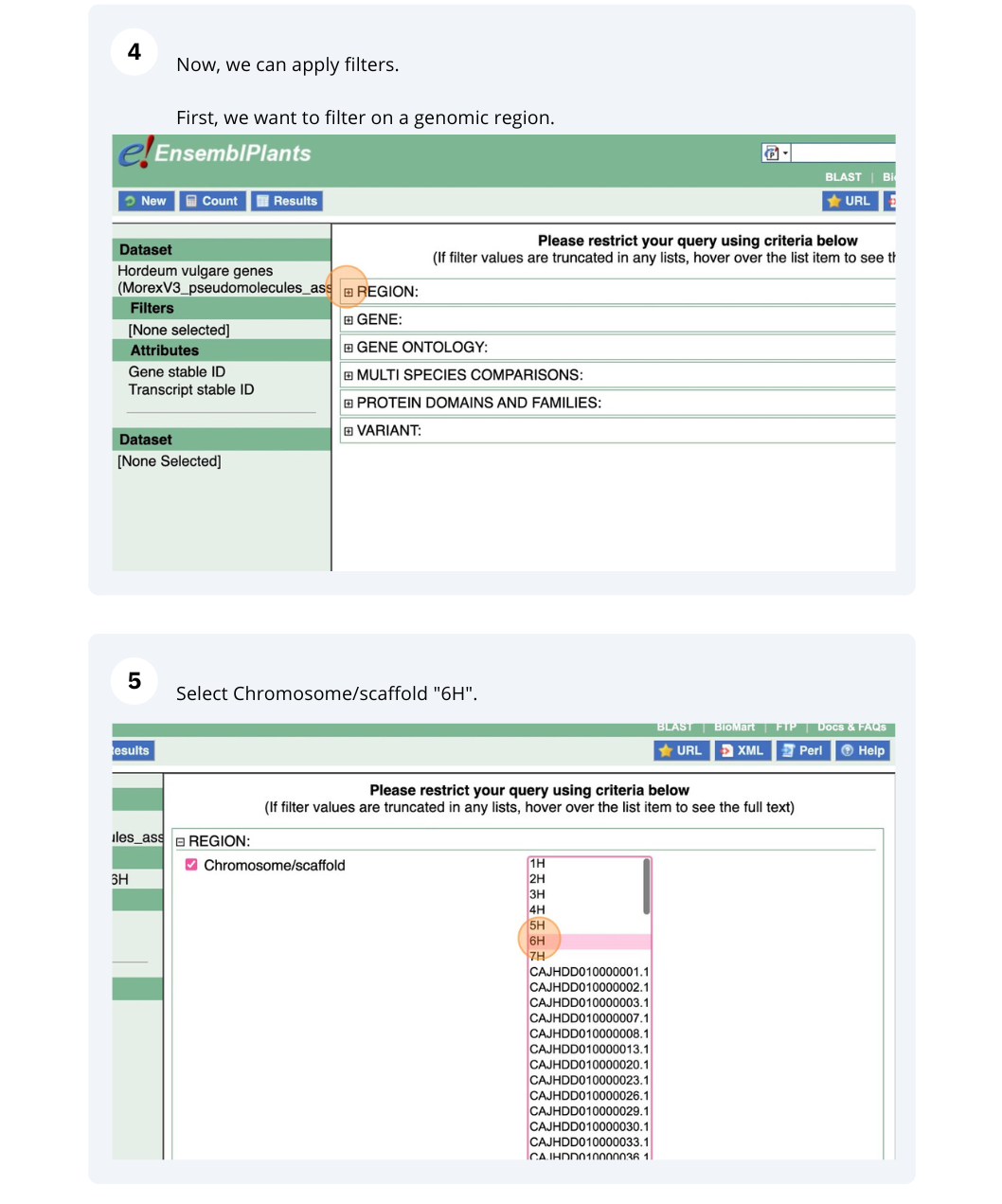
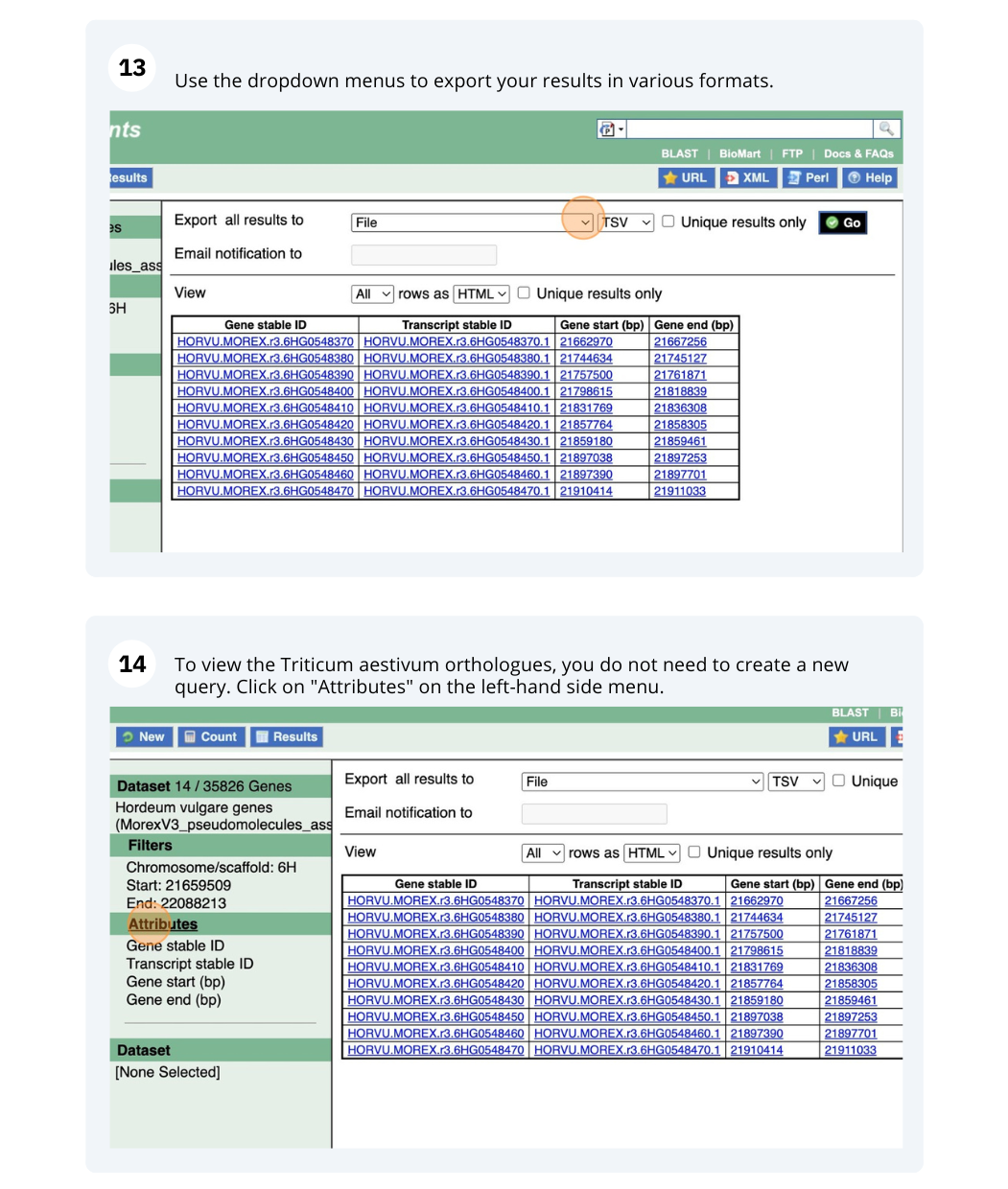
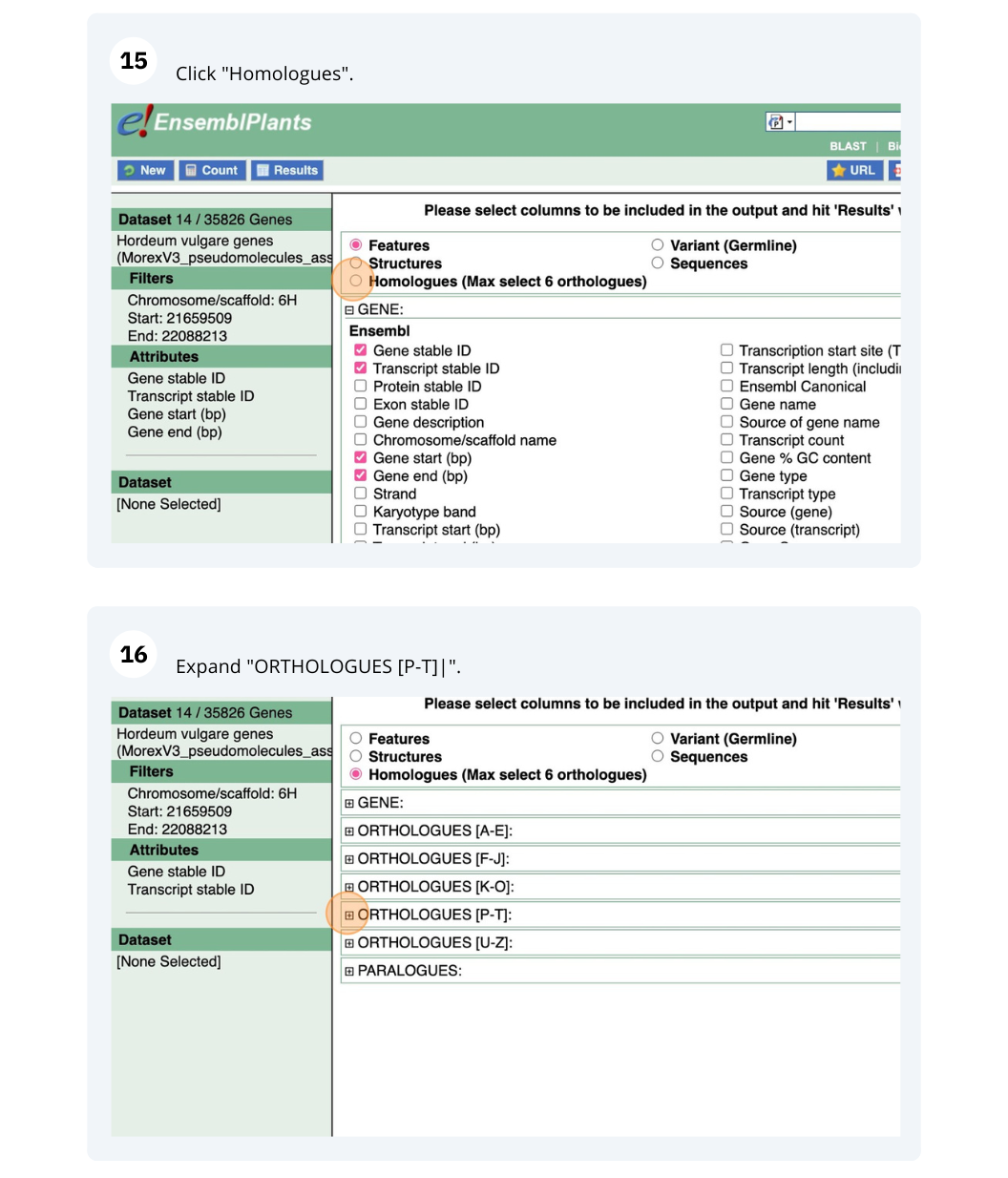
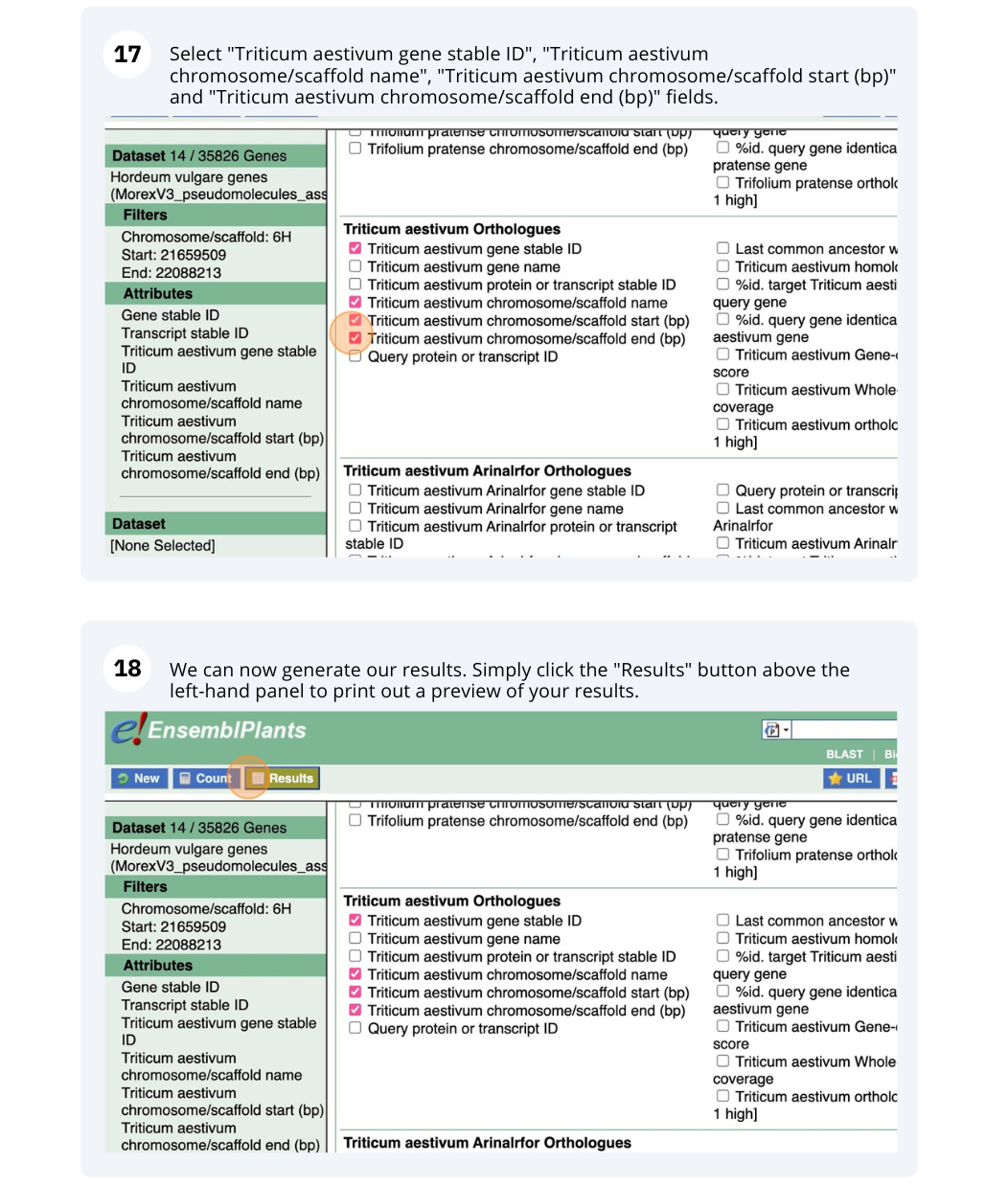
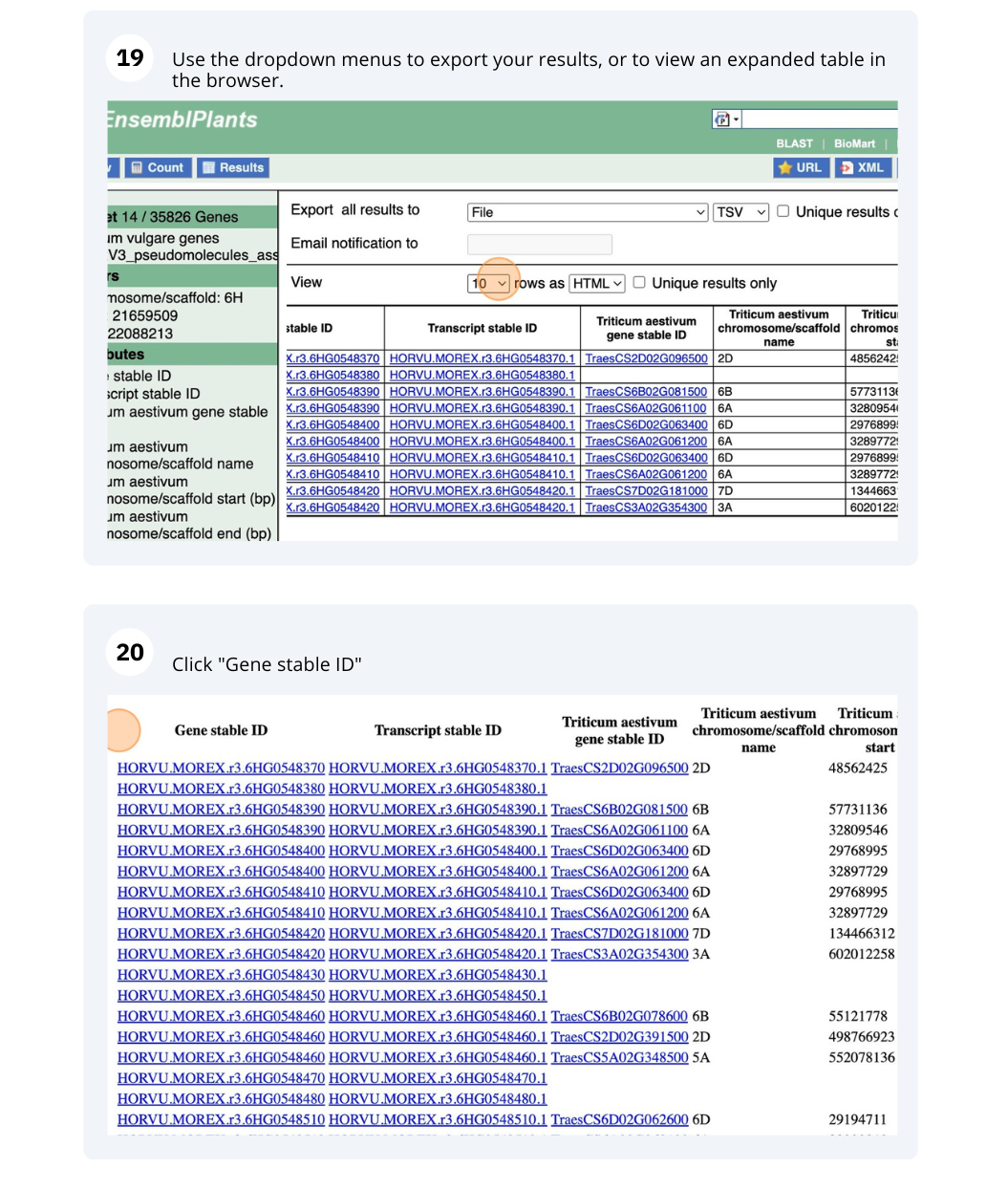
We are interested in finding Hordeum vulgare (MorexV3_pseudomolecules_assembly) genes within a specific region (chromosome 6H, between coordinates 21659509-22088213) and the Triticum aestivum orthologues and their genomic location.
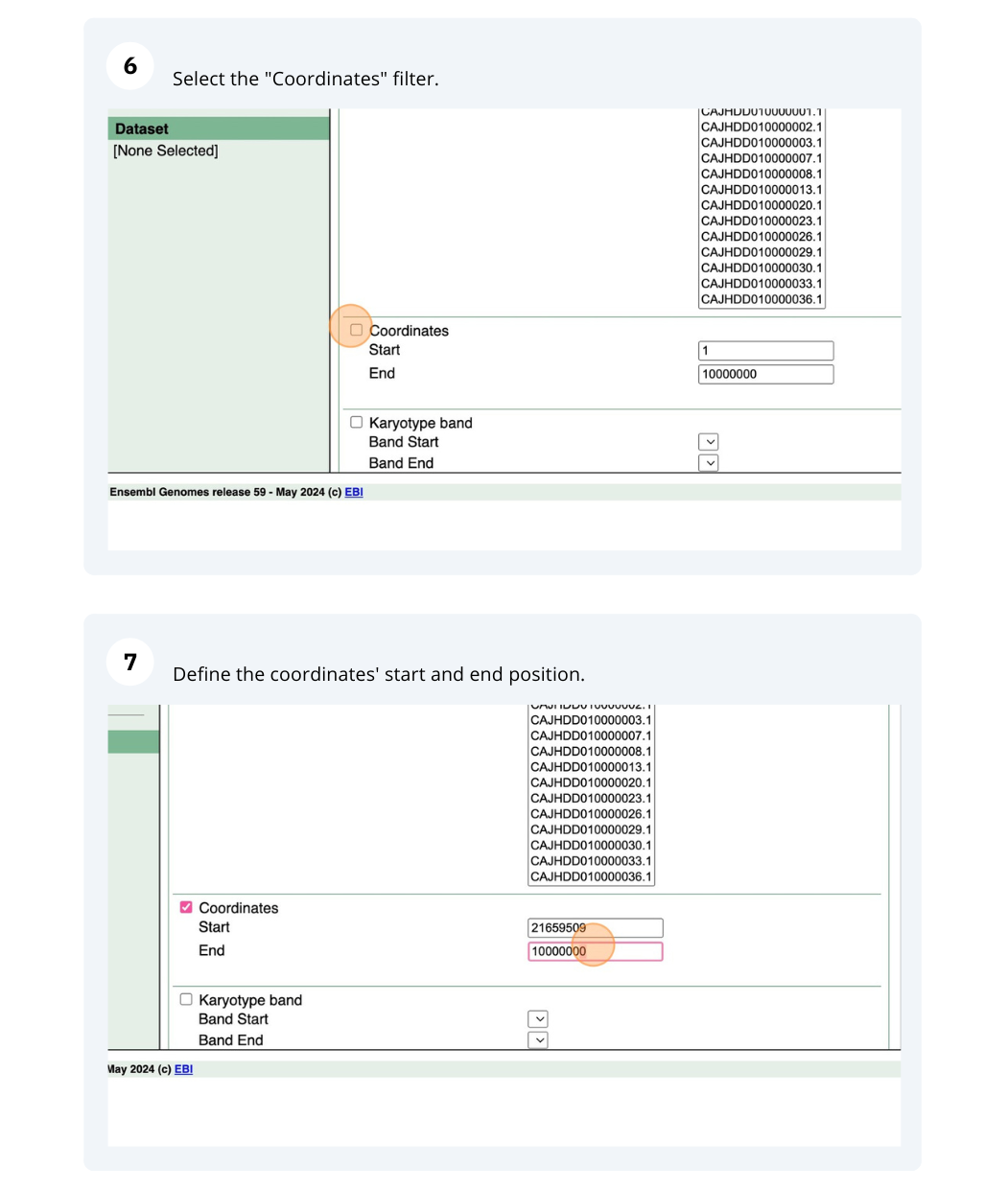
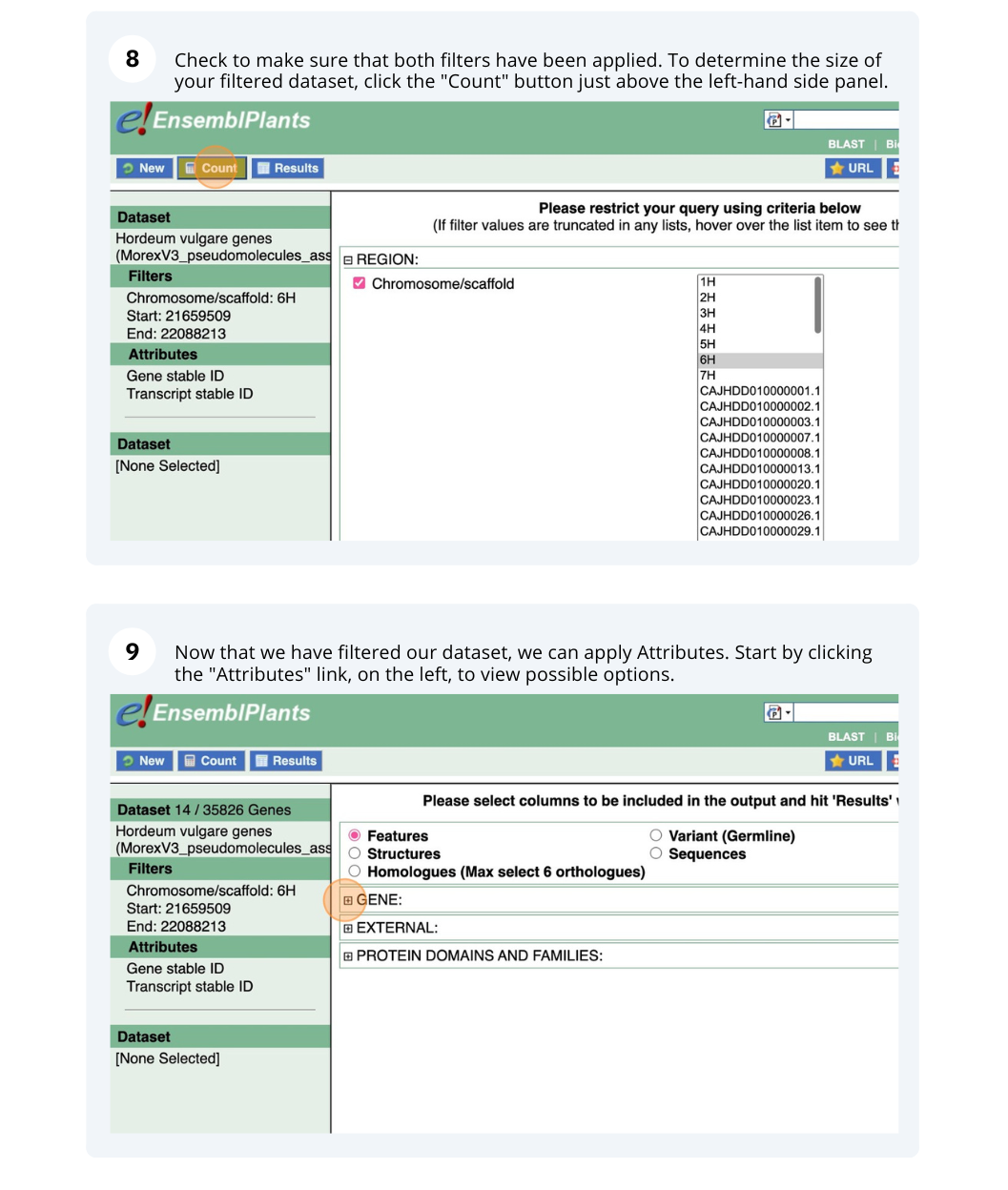
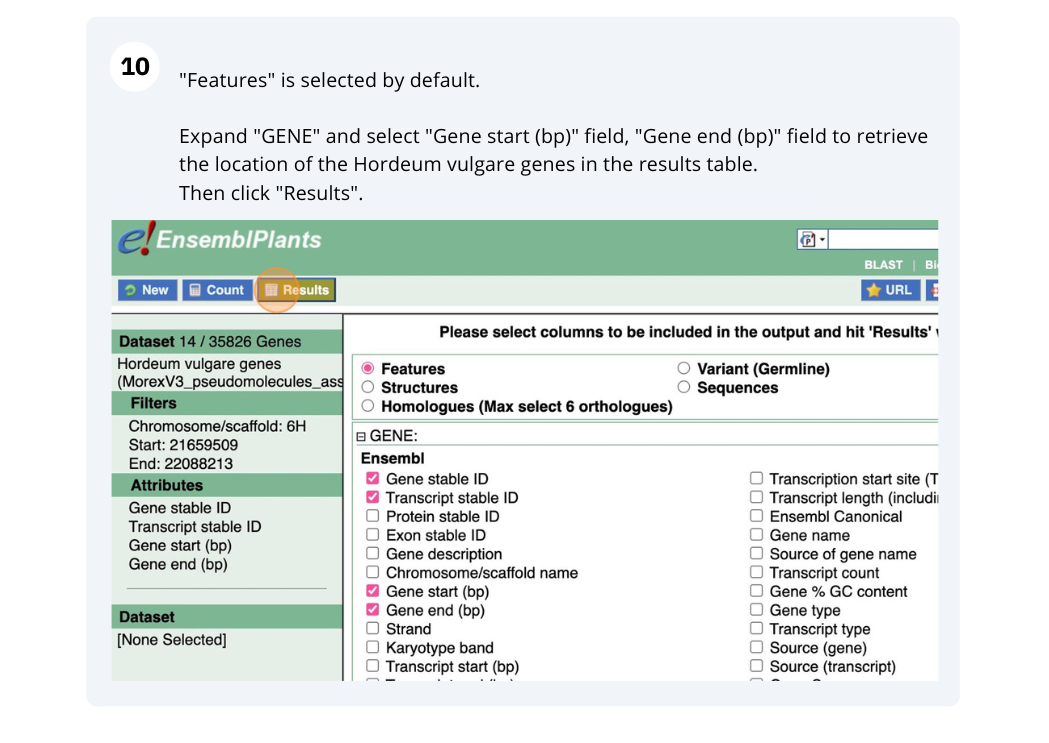
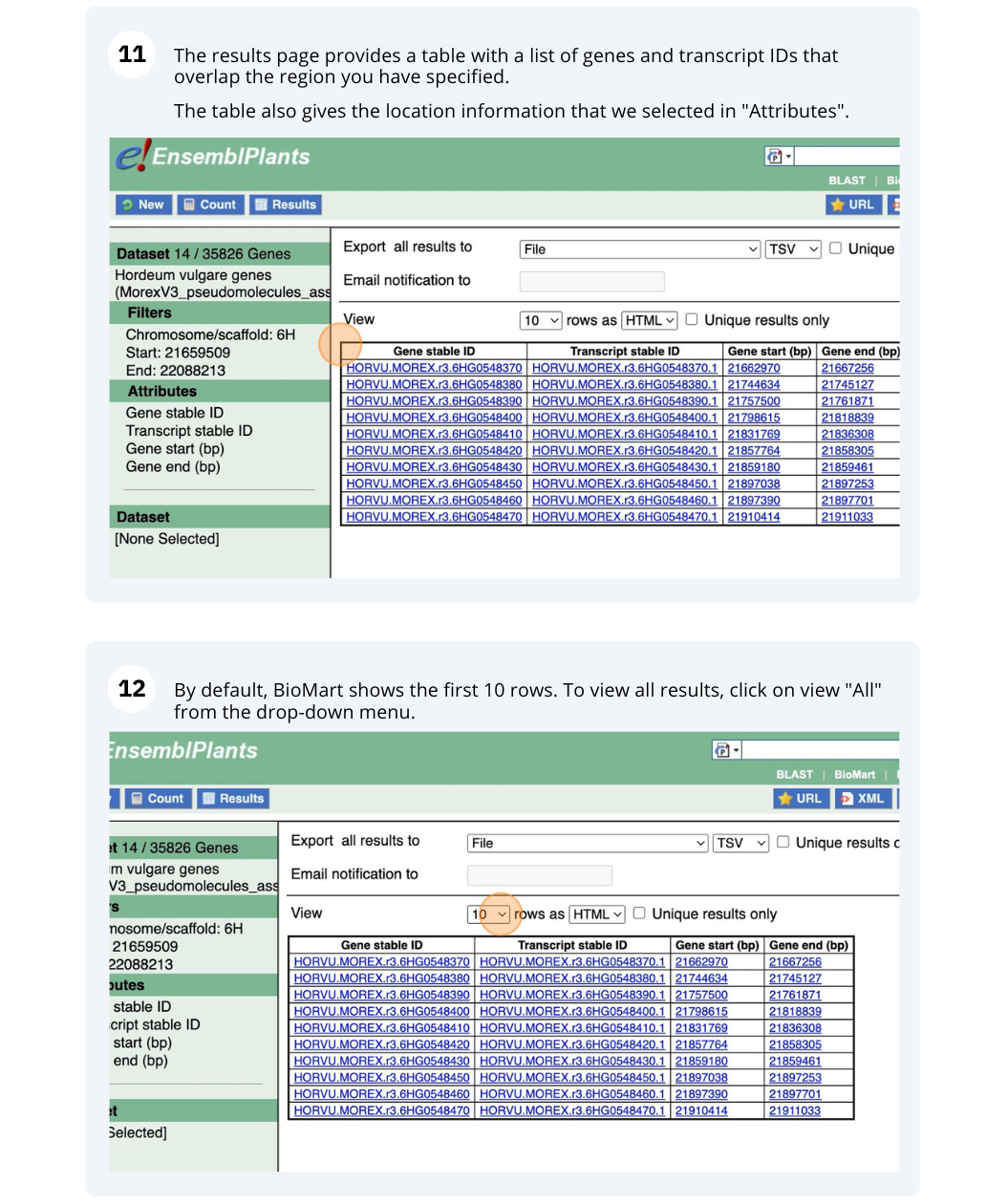
Finding protein coding genes with AlphaFold DB import data in Bemisia tabaci
The whitefly Bemisia tabaci Uganda 1 has been reported from a range of vegetable and weed hosts. This species has been known to transmit different groups of plant-viruses that constrain sweetpotato production in Uganda (Fiallo-Olivé et al. 2020) and a comprehensive understanding of this species is crucial to food security.
- Use BioMart to export a list of protein coding genes in Bemisia tabaci Uganda 1 with AlphaFold DB data
- Retrieve their protein IDs
- Retrieve their sequence in the FASTA format
Go to Ensembl Metazoa. Click on BioMart on the navigation bar at the top of the page. Click the New button on the toolbar on the top left-hand corner, choose the Ensembl Metazoa Genes database and Bemisia tabaci Uganda 1 dataset. Now, filter for the genes with Gene type: Protein coding and Limit to genes: With AlphaFold DB import only.
Make sure the box next to the filter is ticked, otherwise the filter won’t work. Click the Count button on the toolbar.
> This will give you 20 / 13802 Genes.
Go to Attributes on the left-hand panel. Select Gene stable ID, Protein stable ID, AlphaFold DB import Click on Results on the toolbar and the table will display the options you have selected as attributes.
Go to Attributes on the left-hand panel. Expand the SEQUENCES section by clicking on the + box and select Peptide. Select the appropriate header information from the HEADER INFORMATION.
Click on Results on the toolbar and the sequence will be shown as FASTA format. You can export the sequence by downloading it directly to your local machine or sending it to your email.







