Filter Events by Year
Ensembl Metazoa Browser Workshop - Universidad Santiago de Cali
Course Details
- Lead Trainer
- Aleena Mushtaq
- Event Dates
- 2024-07-08 until 2024-07-09
- Location
- Cali, Colombia
- Description
- Work with the Ensembl Outreach team to get to grips with the Ensembl Metazoa browser, accessing gene, variation and comparative genomics data.
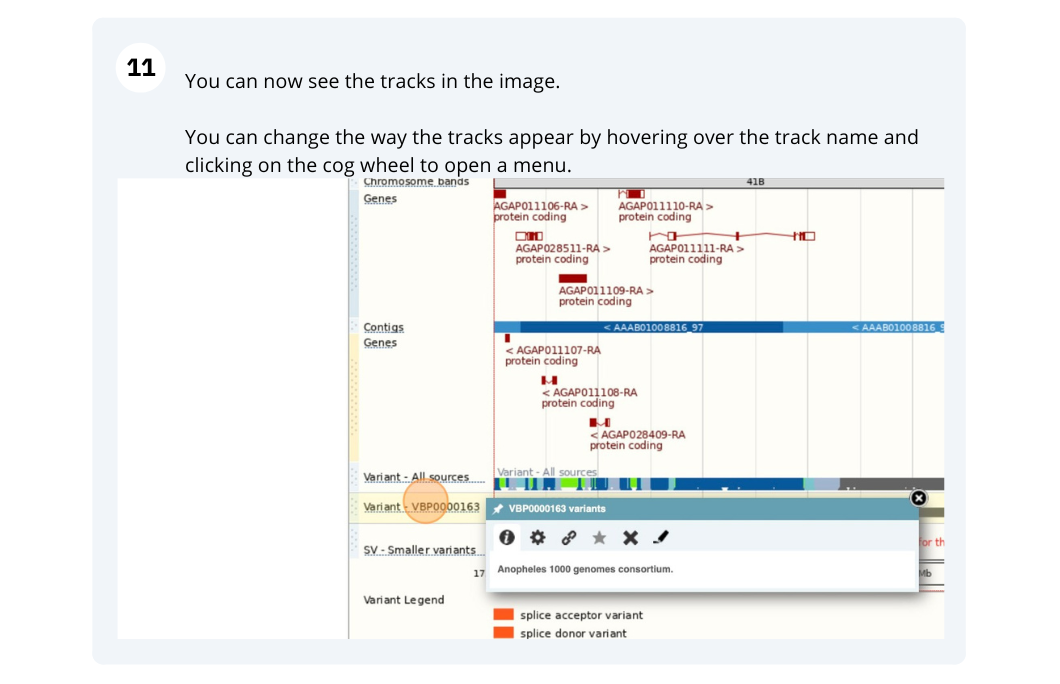
- Survey
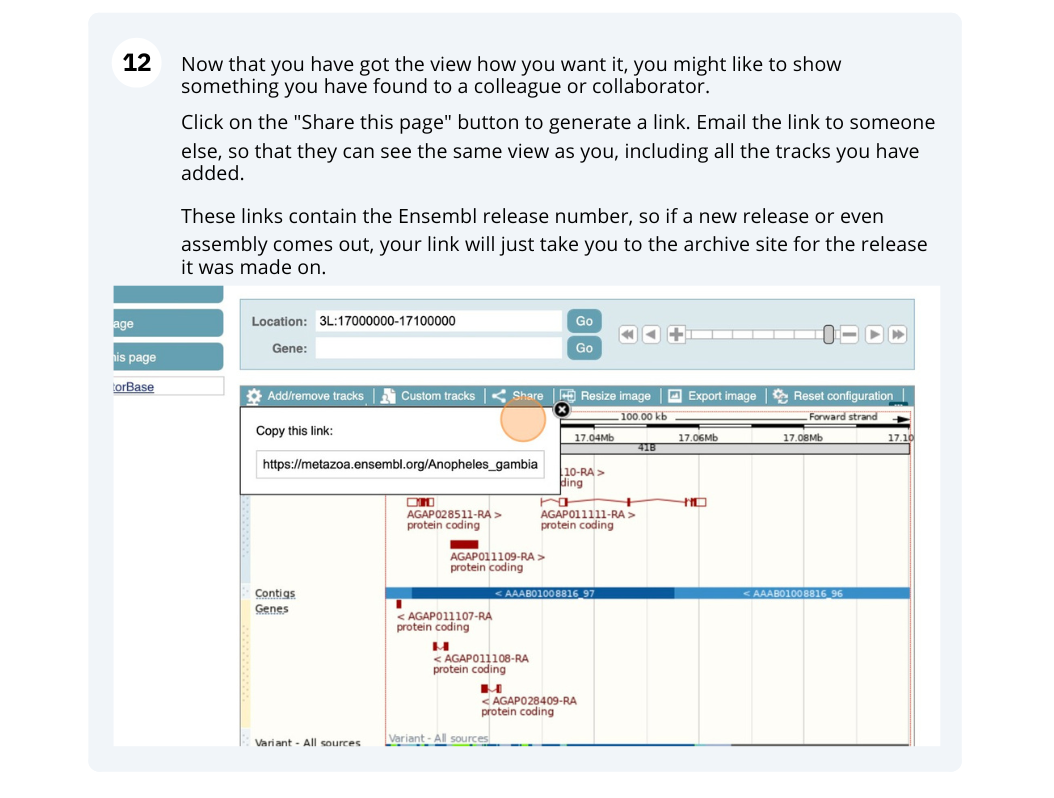
- Ensembl Metazoa Browser Workshop - Universidad Santiago de Cali Feedback Survey
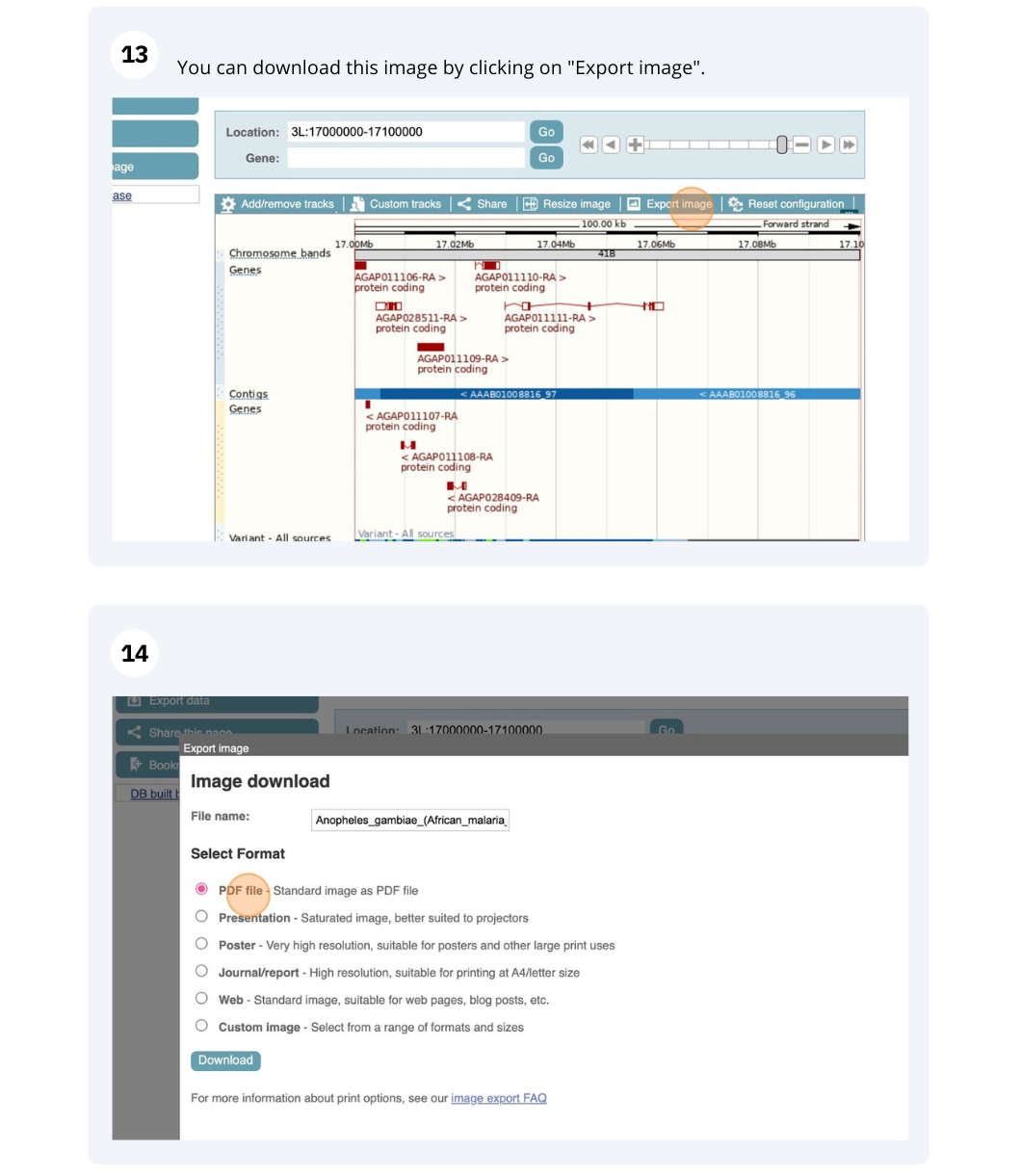
Demos and exercises
Ensembl species
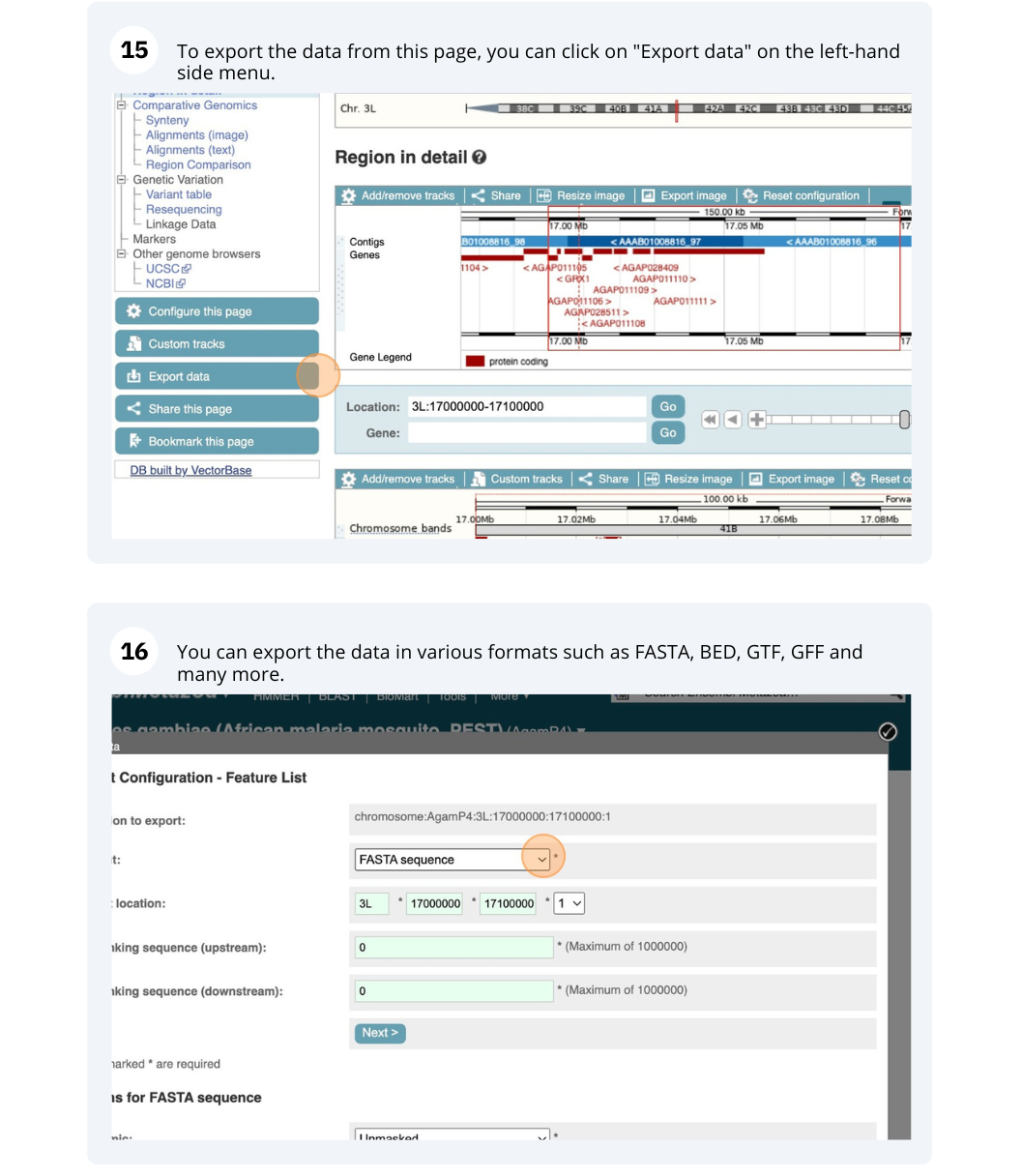
The front page of Ensembl Metazoa is found at www.metazoa.ensembl.org/. It contains lots of information and links to help you navigate Ensembl Metazoa.
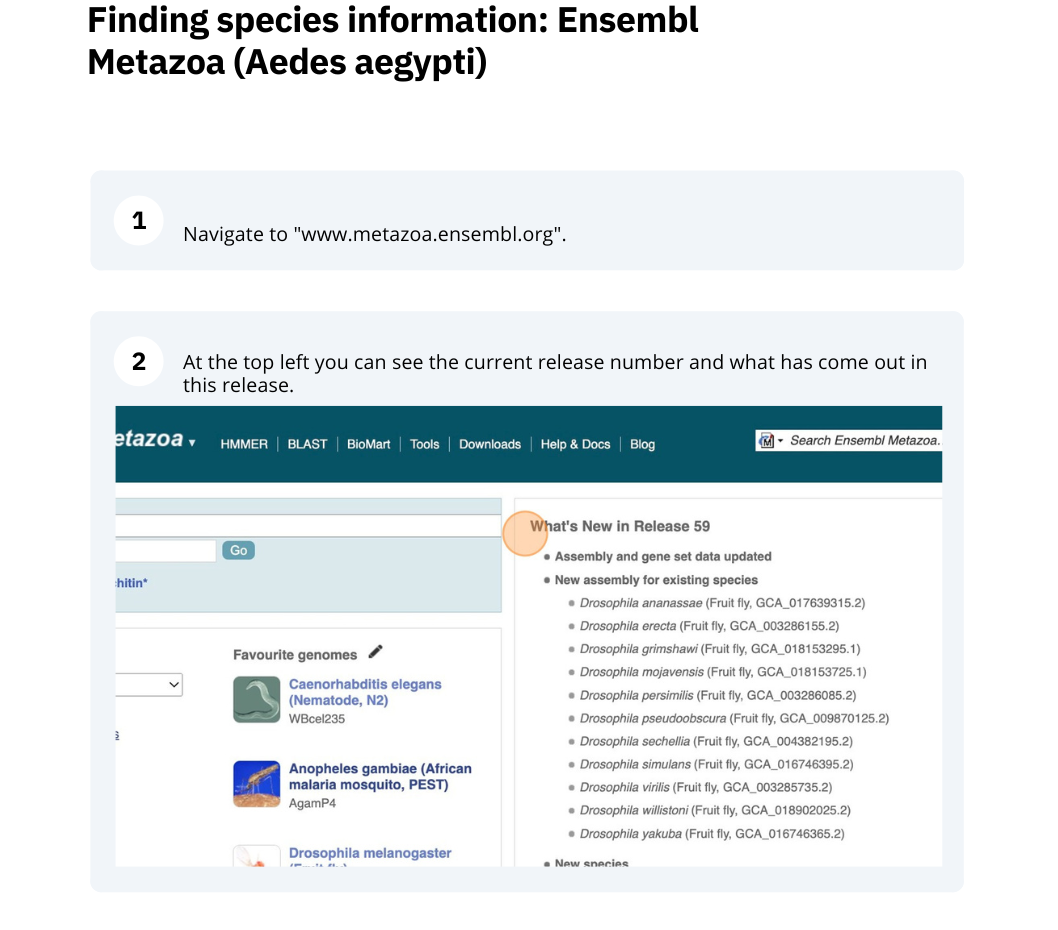
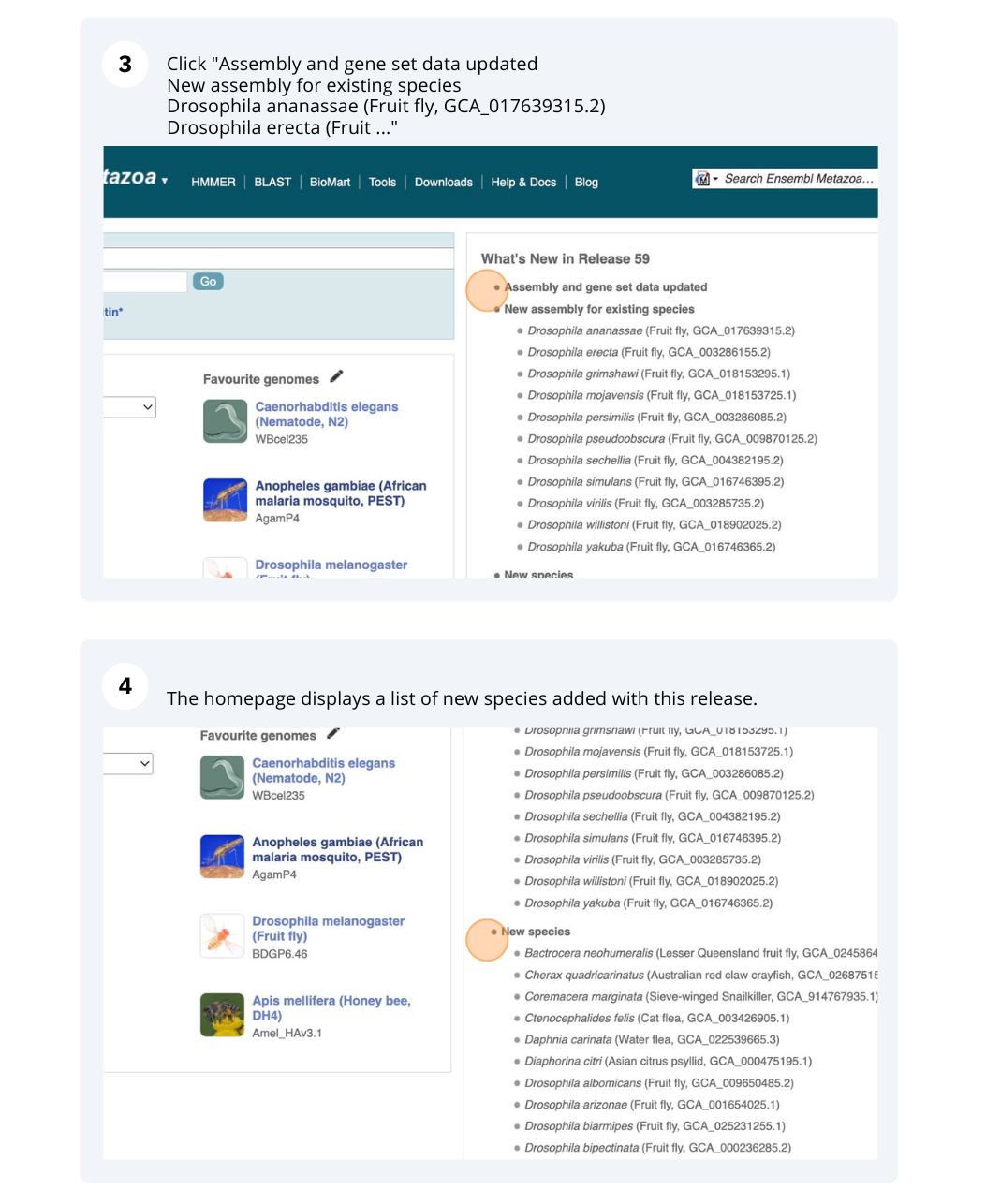
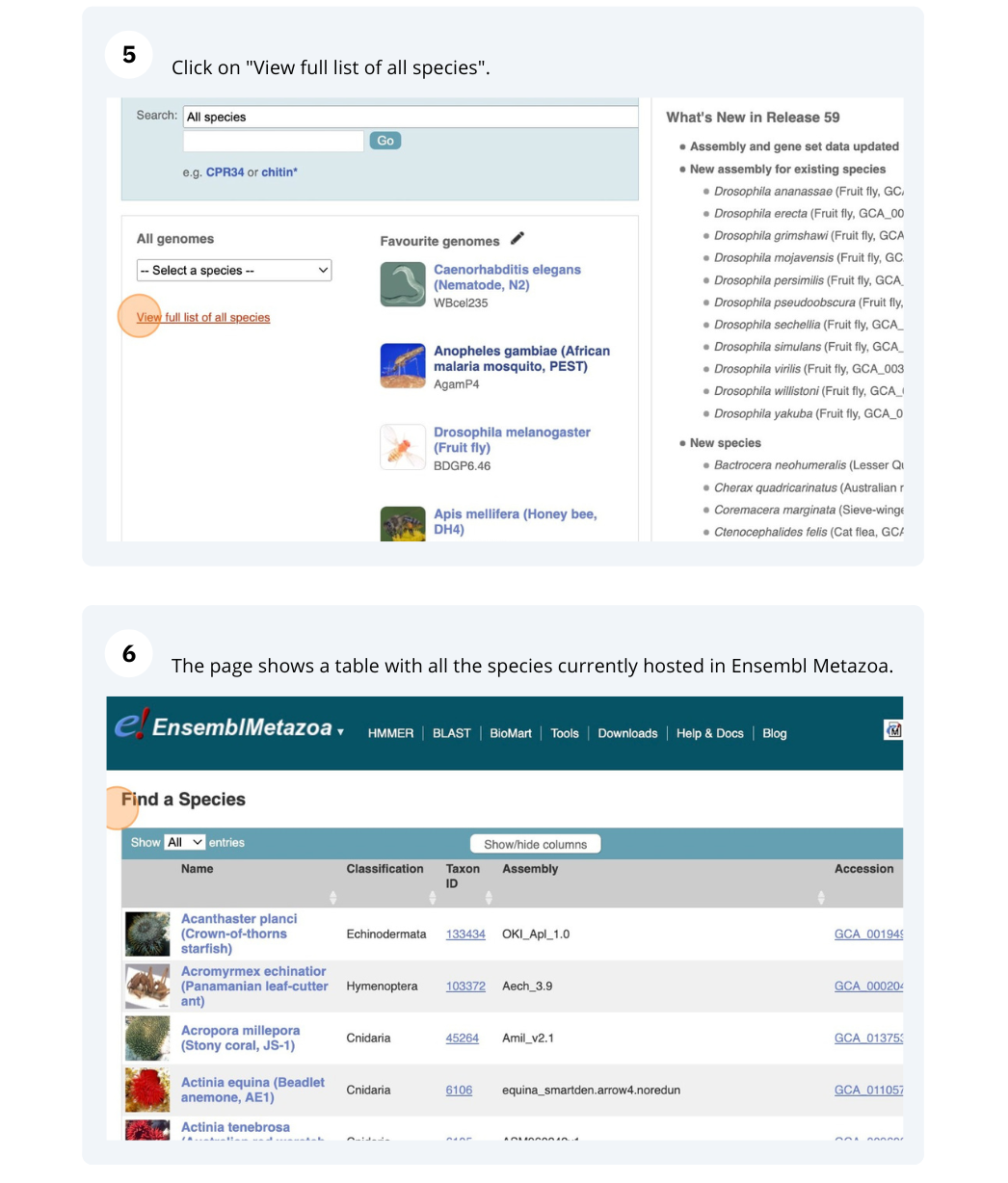
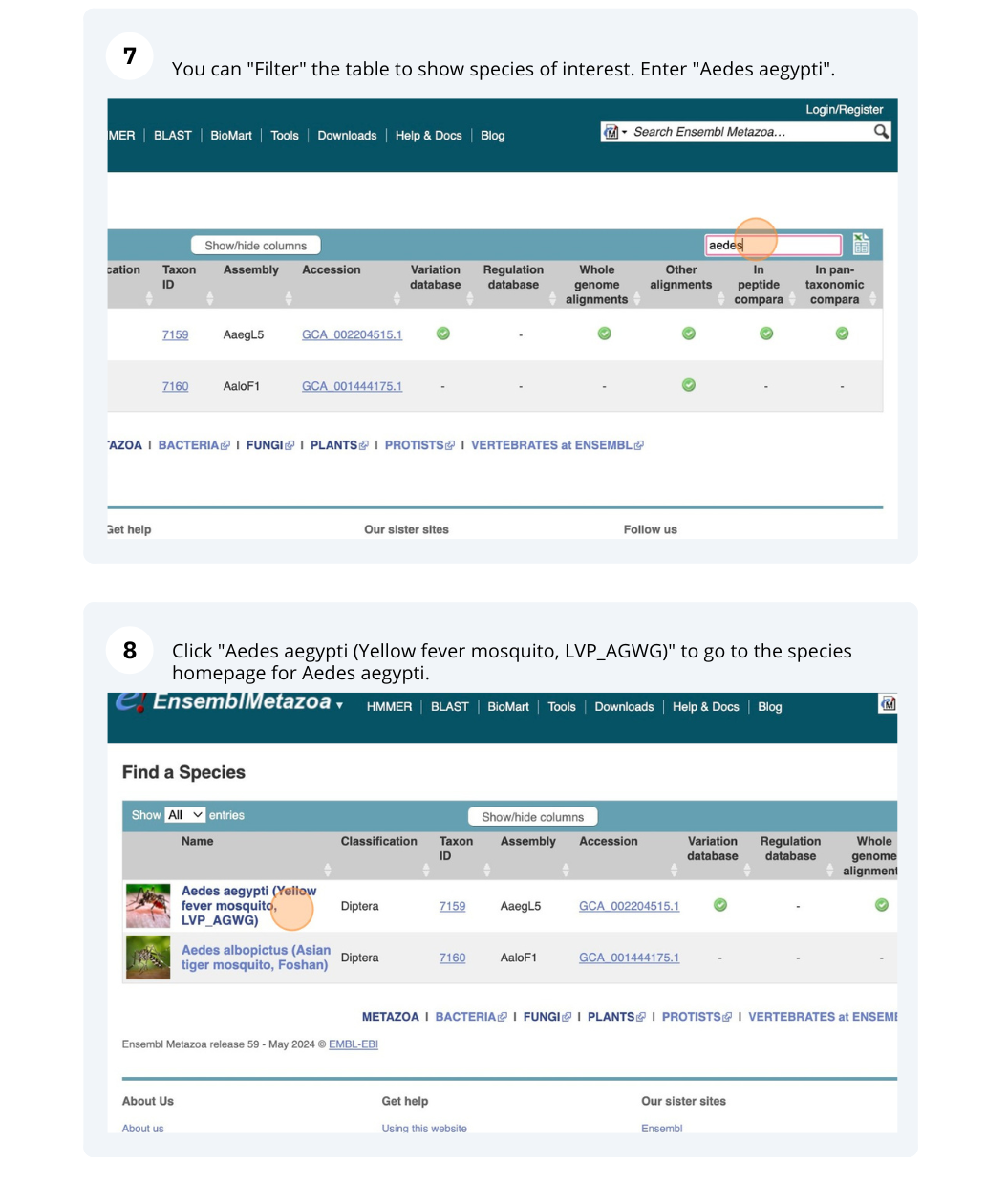
Mosquito species
-
Go to Ensembl Metazoa. How many genomes relating to the genus Anopheles are there in Ensembl Metazoa?
-
When was the current Anopheles gambiae genome assembly last revised?
- Go to metazoa.ensembl.org. Open the drop-down list or click on View full list of all Ensembl Metazoa species. In a latin binomial species name, the first word represents the genus. Type Anopheles into the filter box in the top left to find all genomes with this word in the binomial.
There are 22 Anopheles genomes (some species are represented by more than one genome).
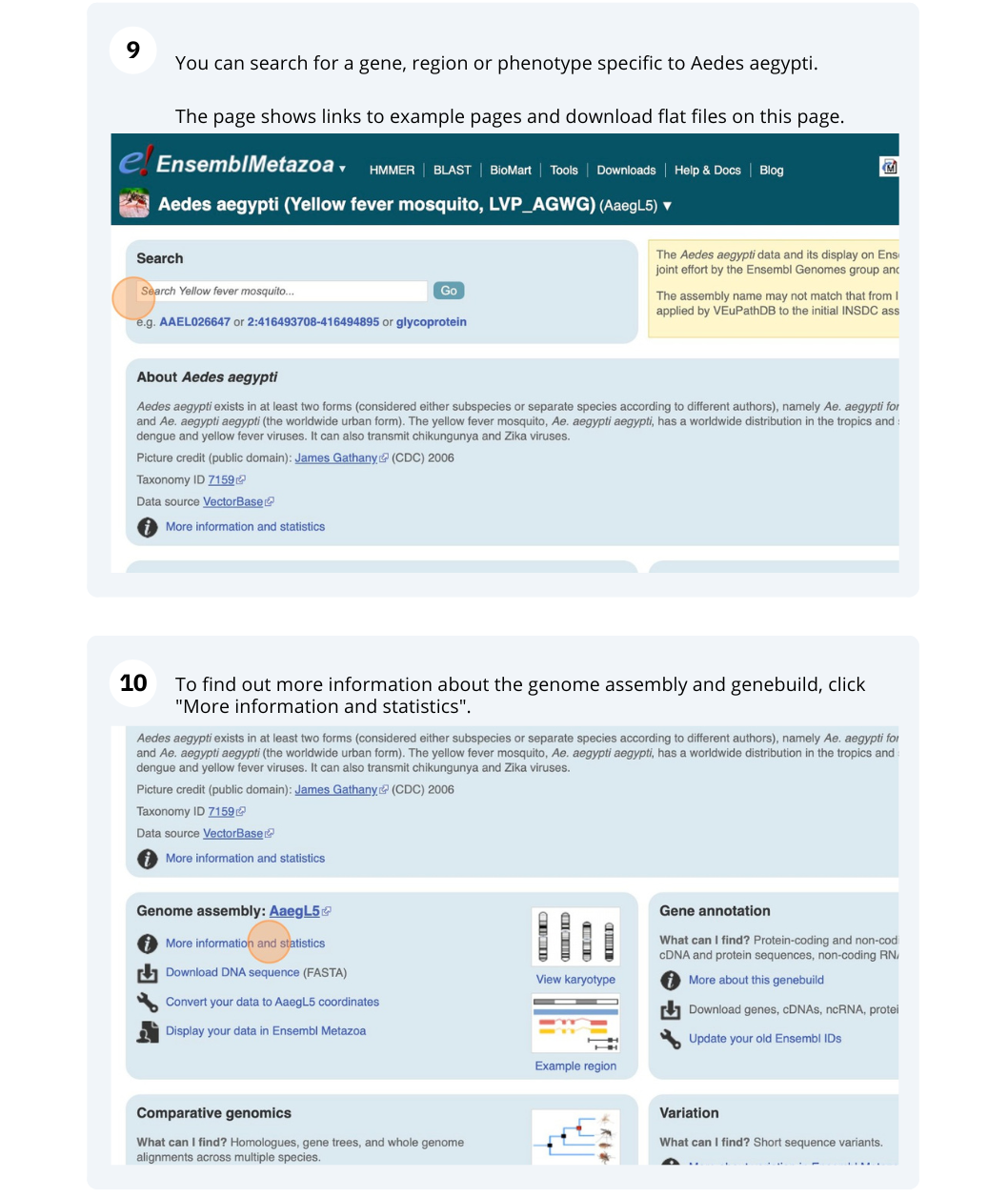
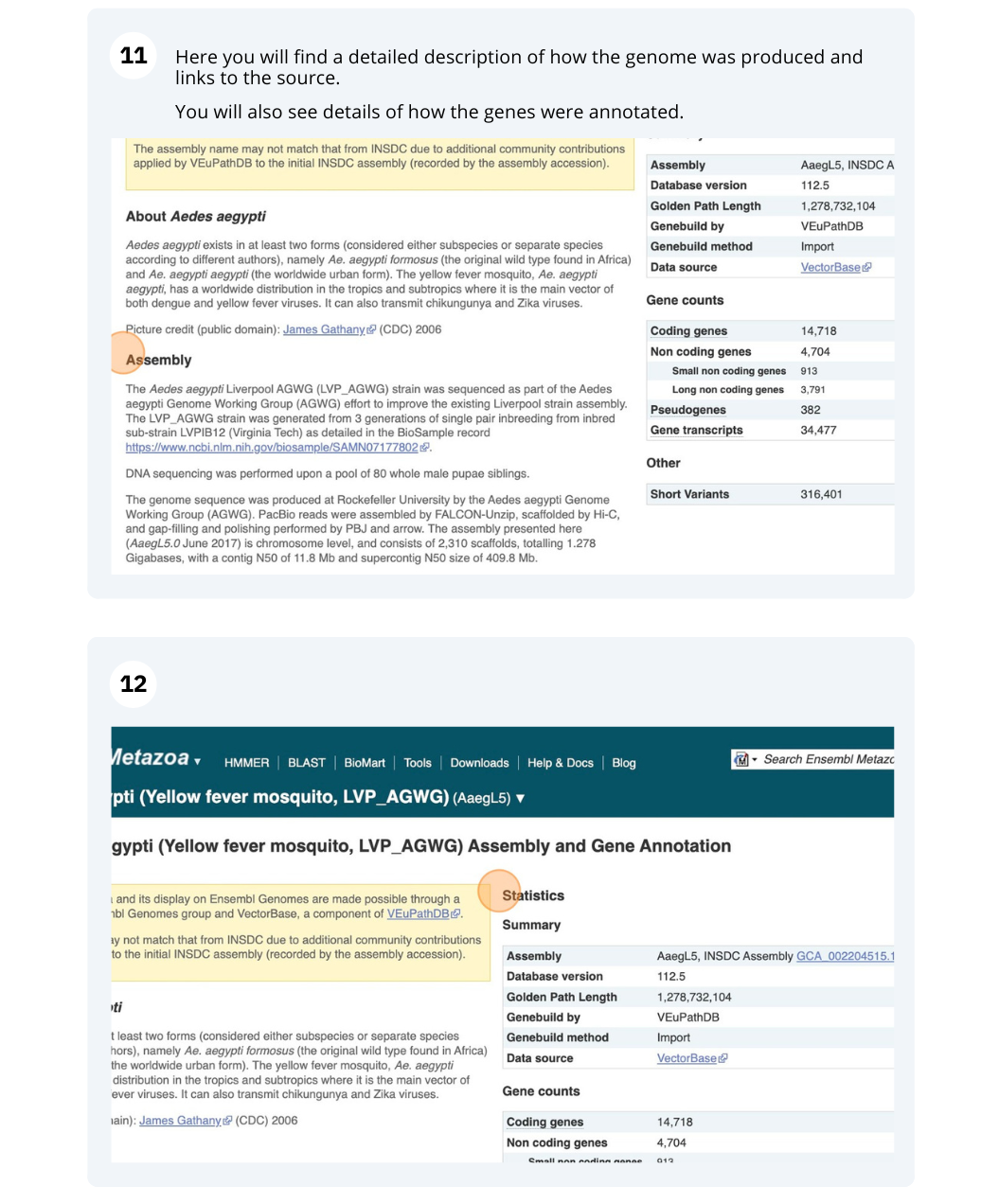
- Click on Anopheles gambiae (African malaria mosquito, PEST), and then on More information and statistics.
The assembly hosted is AgamP4 (INSDC Assembly GCA_000005575.1) which was revised in Feb 2006.
Exploring genomic regions
Demo: Exploring genomic regions in Ensembl Metazoa
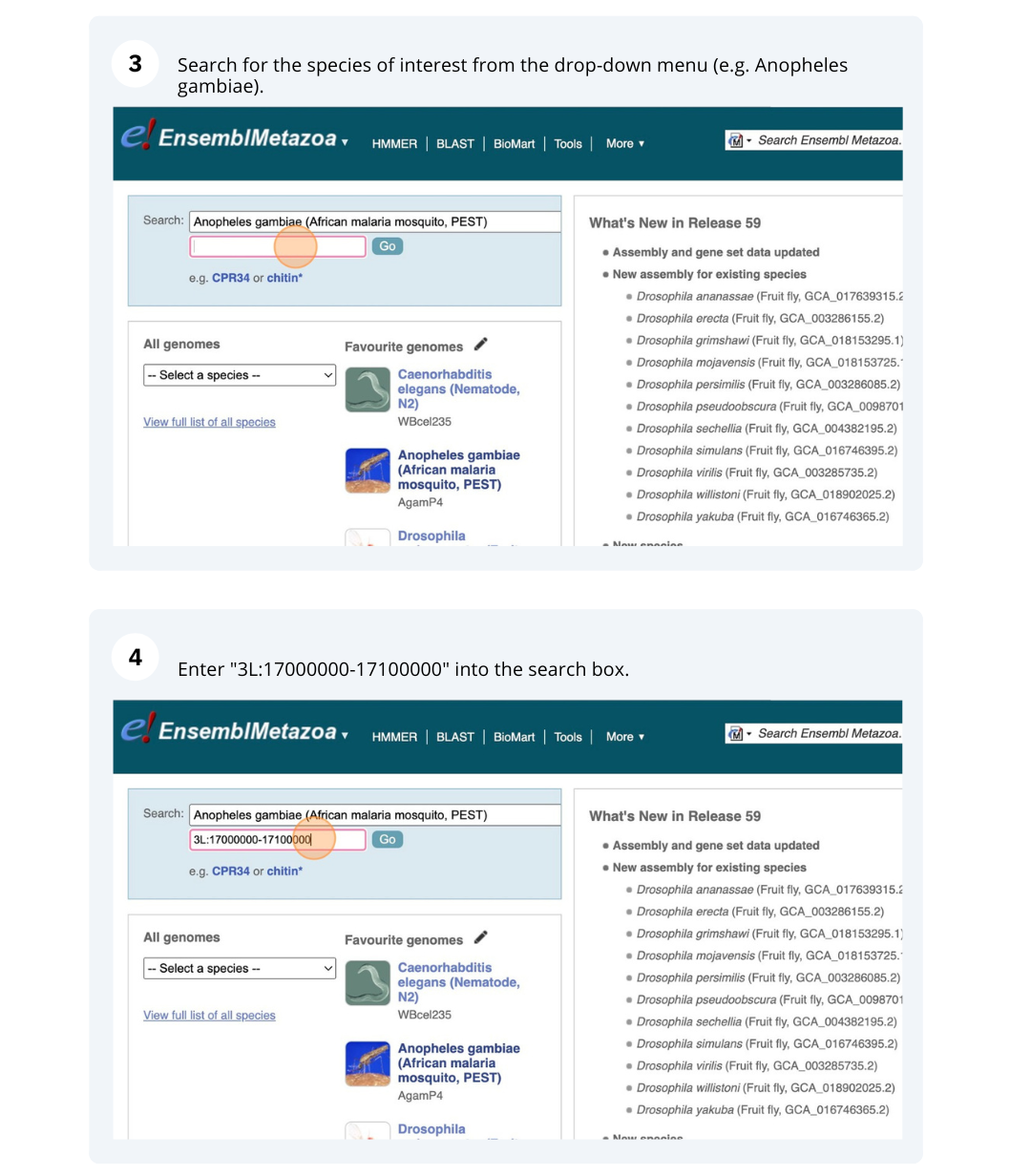
We’re going to look at a region of the Anopheles gambiae (African malaria mosquito, PEST) genome, 3L:17000000-17100000, and manipulate the view to see the data we are interested in.

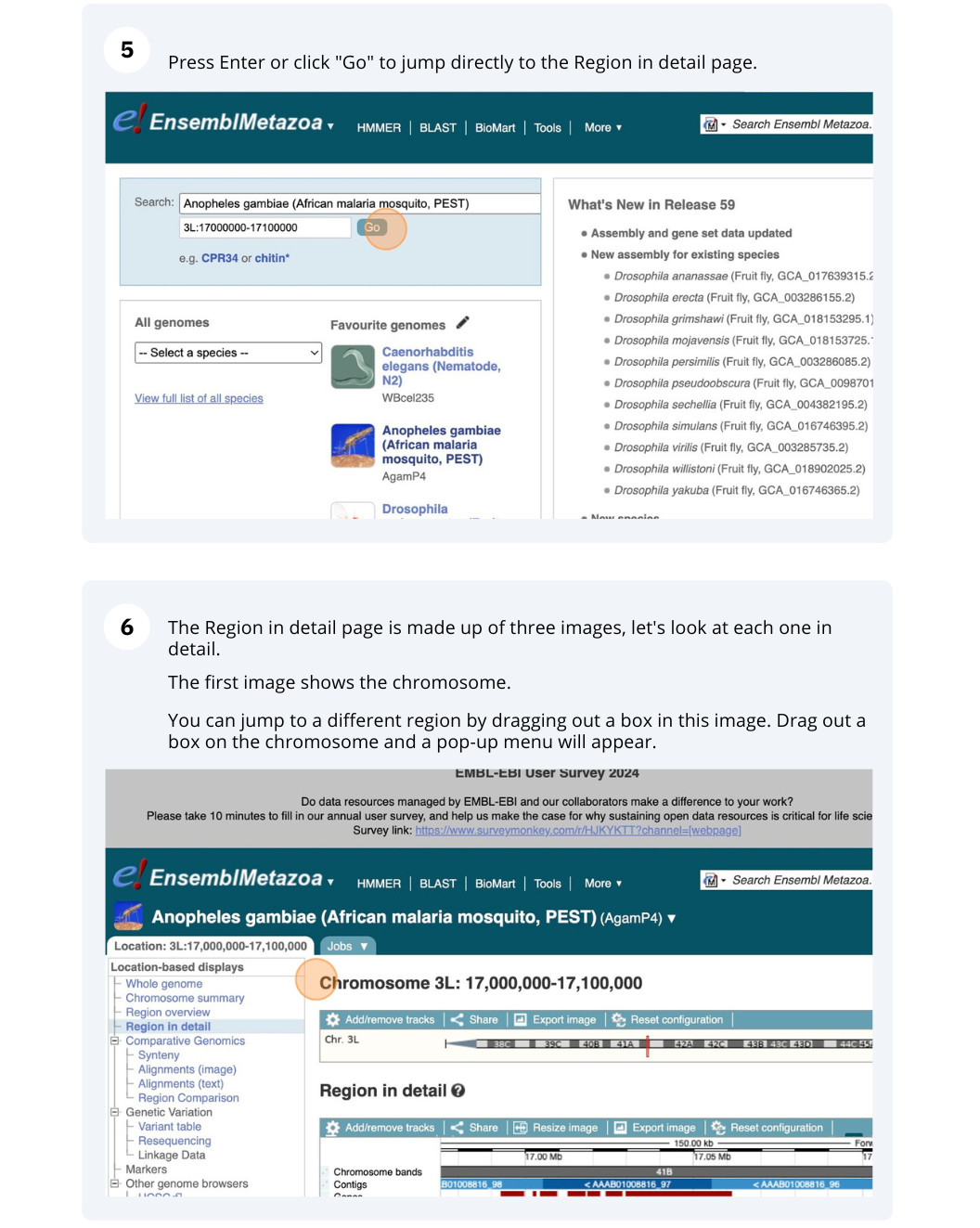
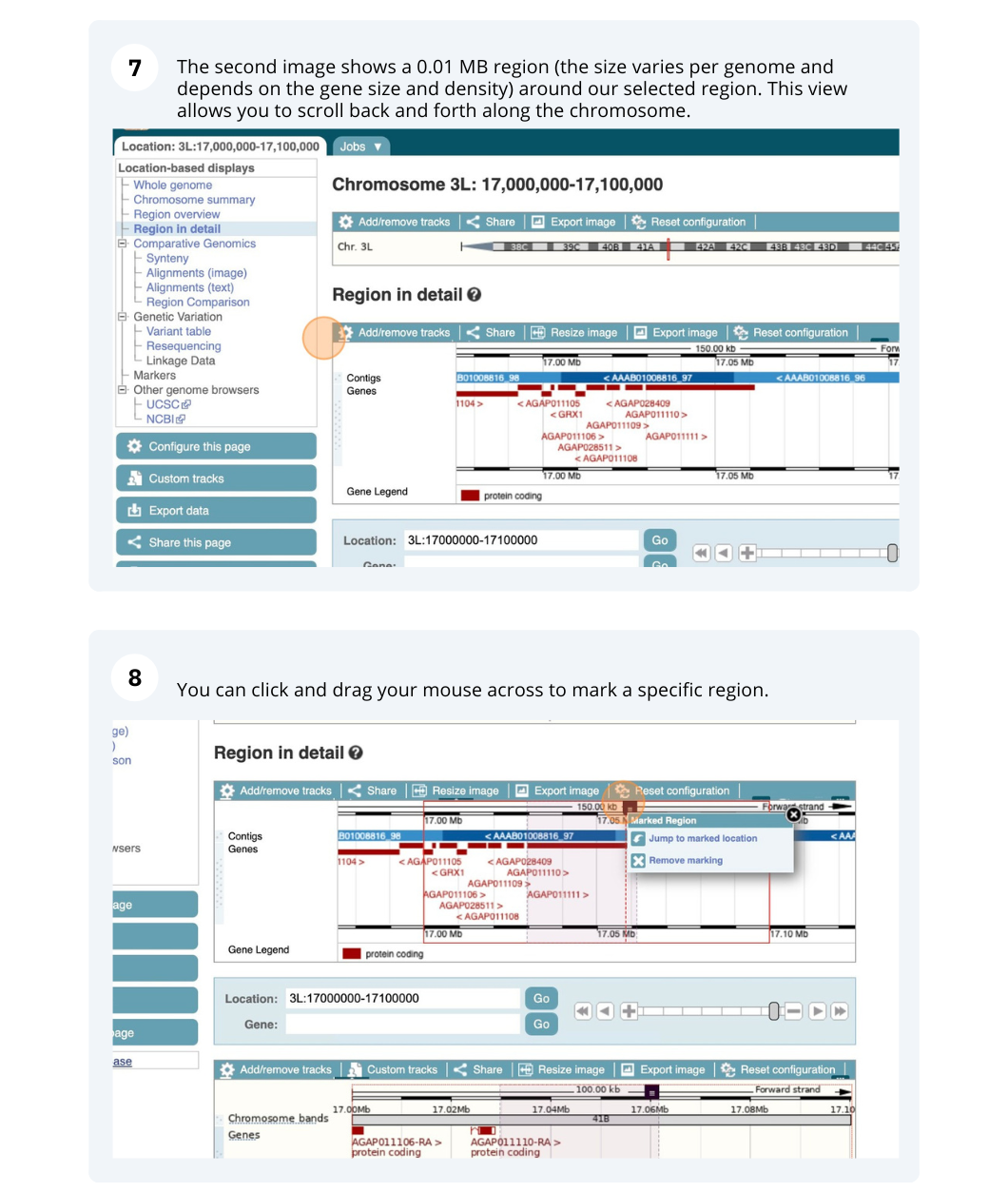
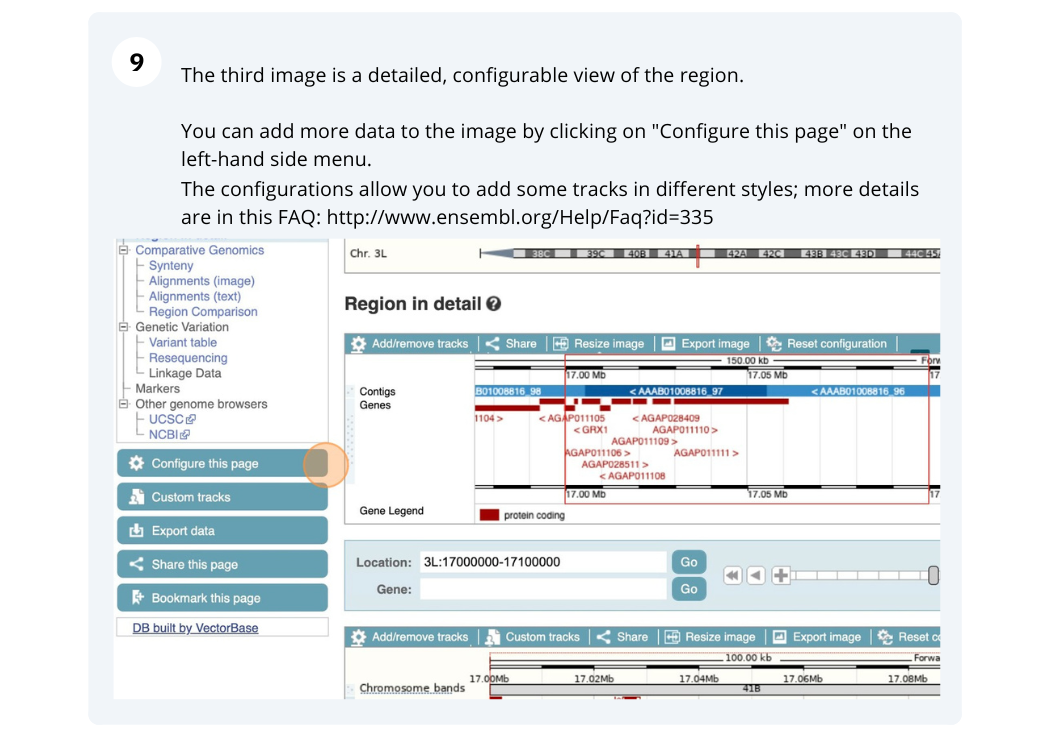
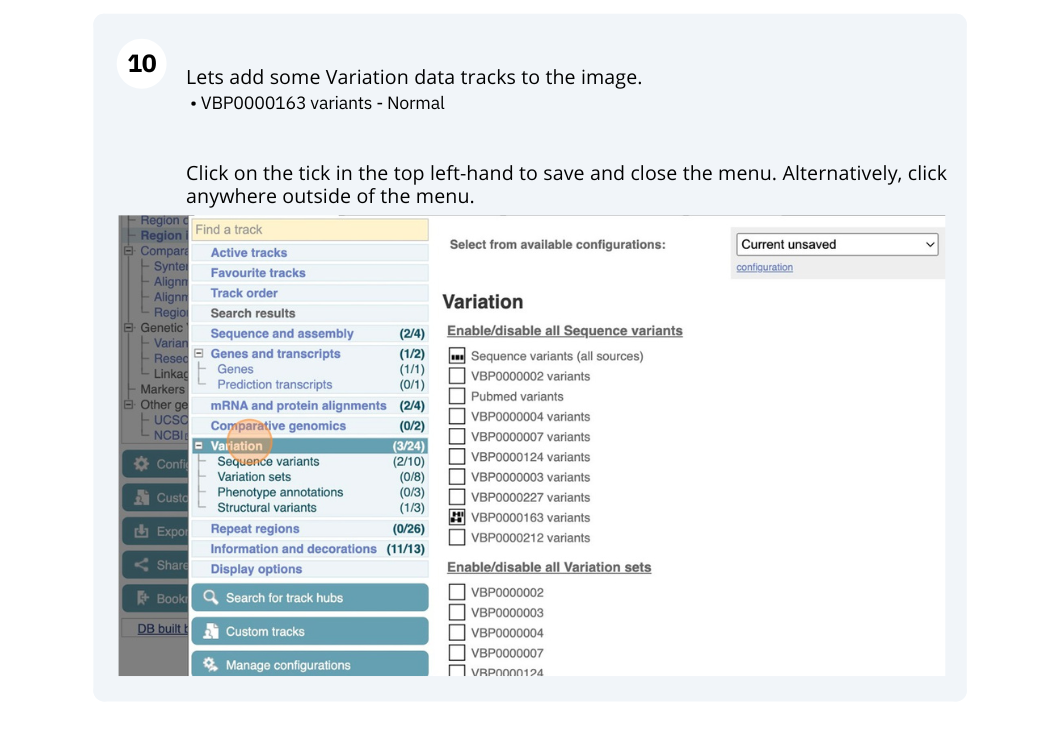
Exploring a genomic region in Anopheles gambiae
(a) Go to the region from 7,300,000 to 7,450,000 bp on Anopheles gambiae chromosome 2L. On which cytogenetic band is this region located?
(b) How many genes are found in this region? Zoom in on the second exon of AGAP004970-RA. Turn on the track Start/Stop codons. Can you see the start codon of AGAP004970-RA?
(c) Highlight the start codon of AGAP004970-RA. Zoom out to view the whole gene. Can you see where you highlighted?
(a) Go to the Ensembl metazoa homepage.
Select Search: Anopheles gambiae (African malaria mosquito, PEST), type 2L:7300000-7450000 and click Go.
The region is located on the cytogenetic band 21B.
(b) There are nine genes within this region and one that overlaps the end.
Drag out a box around the second exon of the gene, the second red box from the left and click on Jump to region in the pop-up window to zoom in. If you have not zoomed in far enough, drag out another box and click on Jump to region.
The nucleotide sequence will appear either side of the blue contig as pale blue (C), yellow (G), green (A) and pink (T) boxes. As you zoom in further, you will see the letters on the bases.
Click on Configure this page and click on Sequence and assembly. Turn on the track for Start/stop codons.
Alternatively, you can find the tracks by typing the name into the yellow Find a track box at the top right. Close the menu.
Start codons are shown in green and stop codons in red. You should see a green start codon that coincides with the start of the filled in red box.
(c) Drag out a box around the green start codon and select Mark region. The highlighted region should be visible as a grey dotted line. Scroll up to the overview to drag out a box around the gene and select Jump to region, you will still see the highlight in this view.
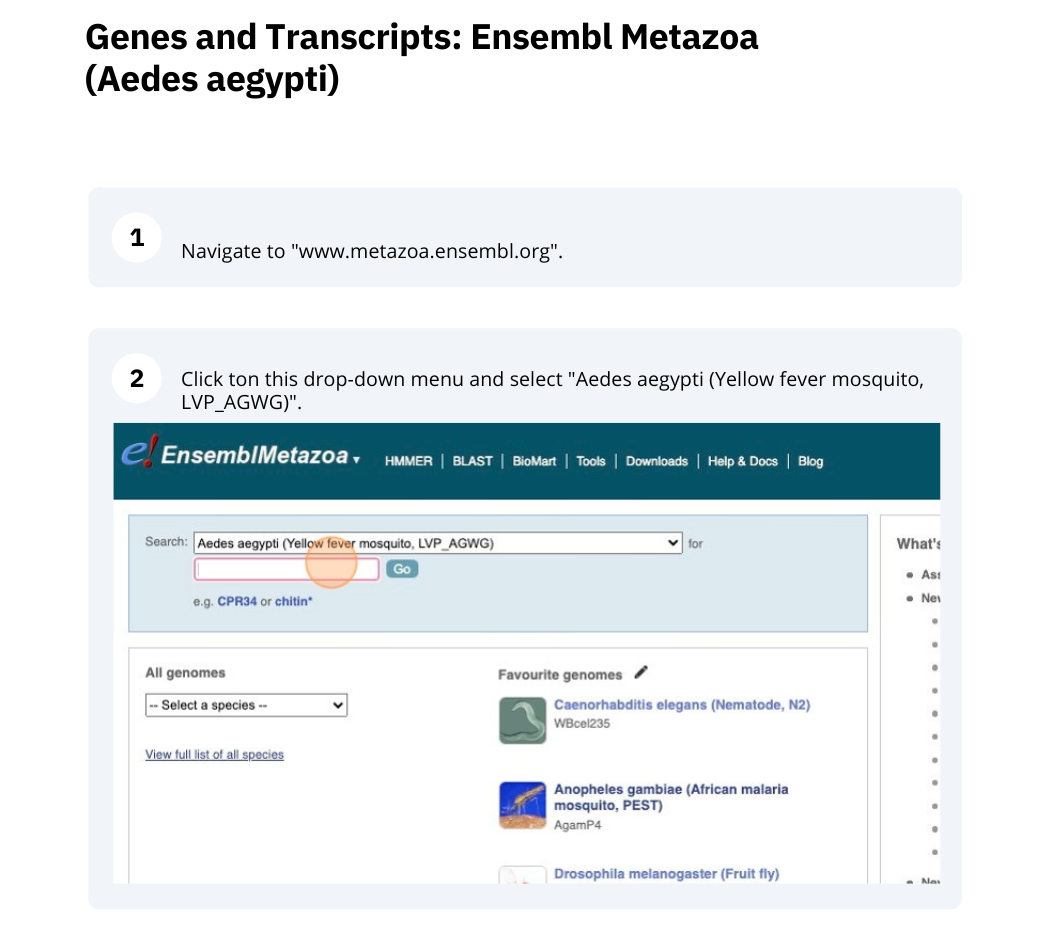
Genes and Transcripts
Demo: Exploring genes and transcripts in Ensembl Metazoa
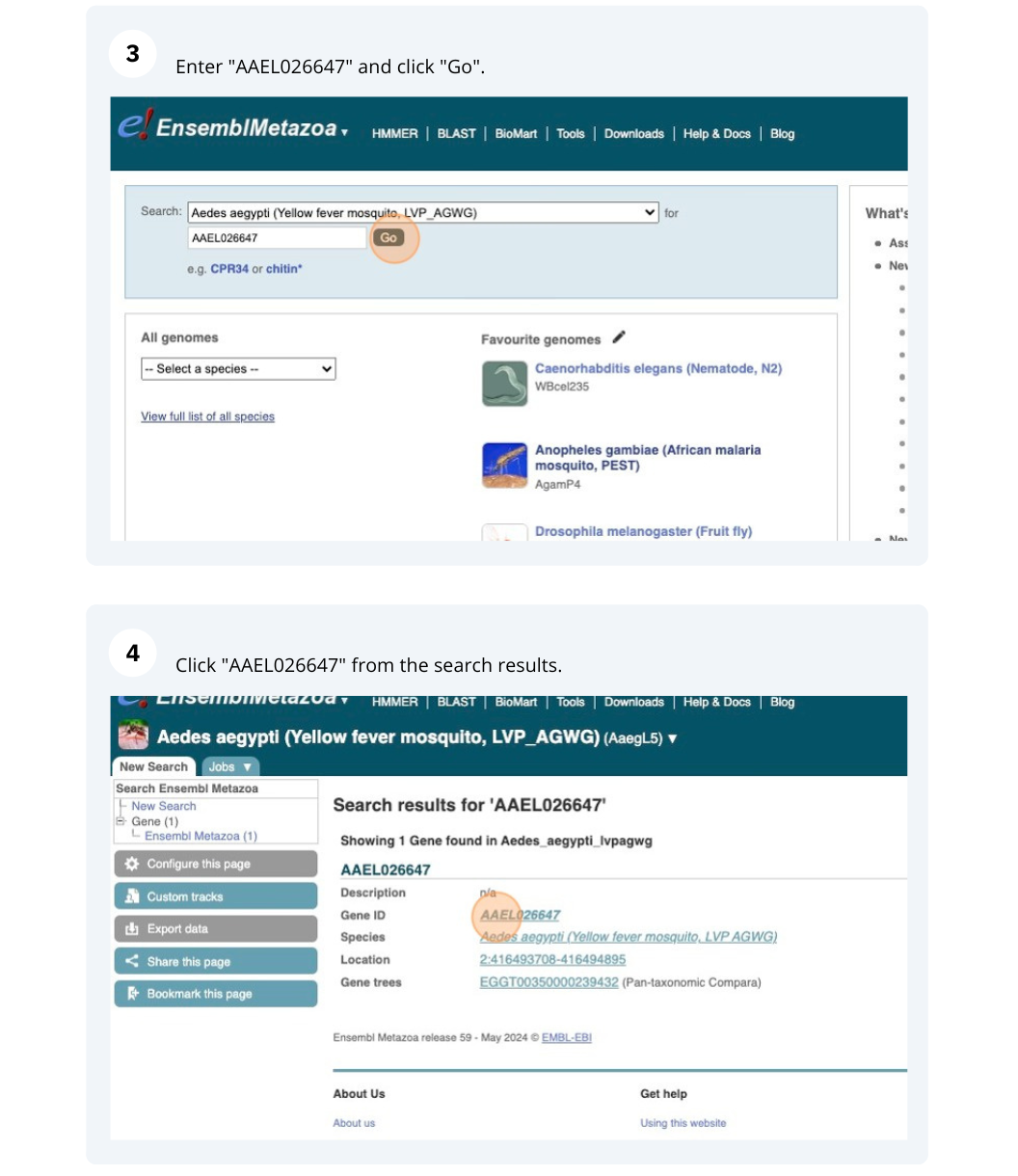
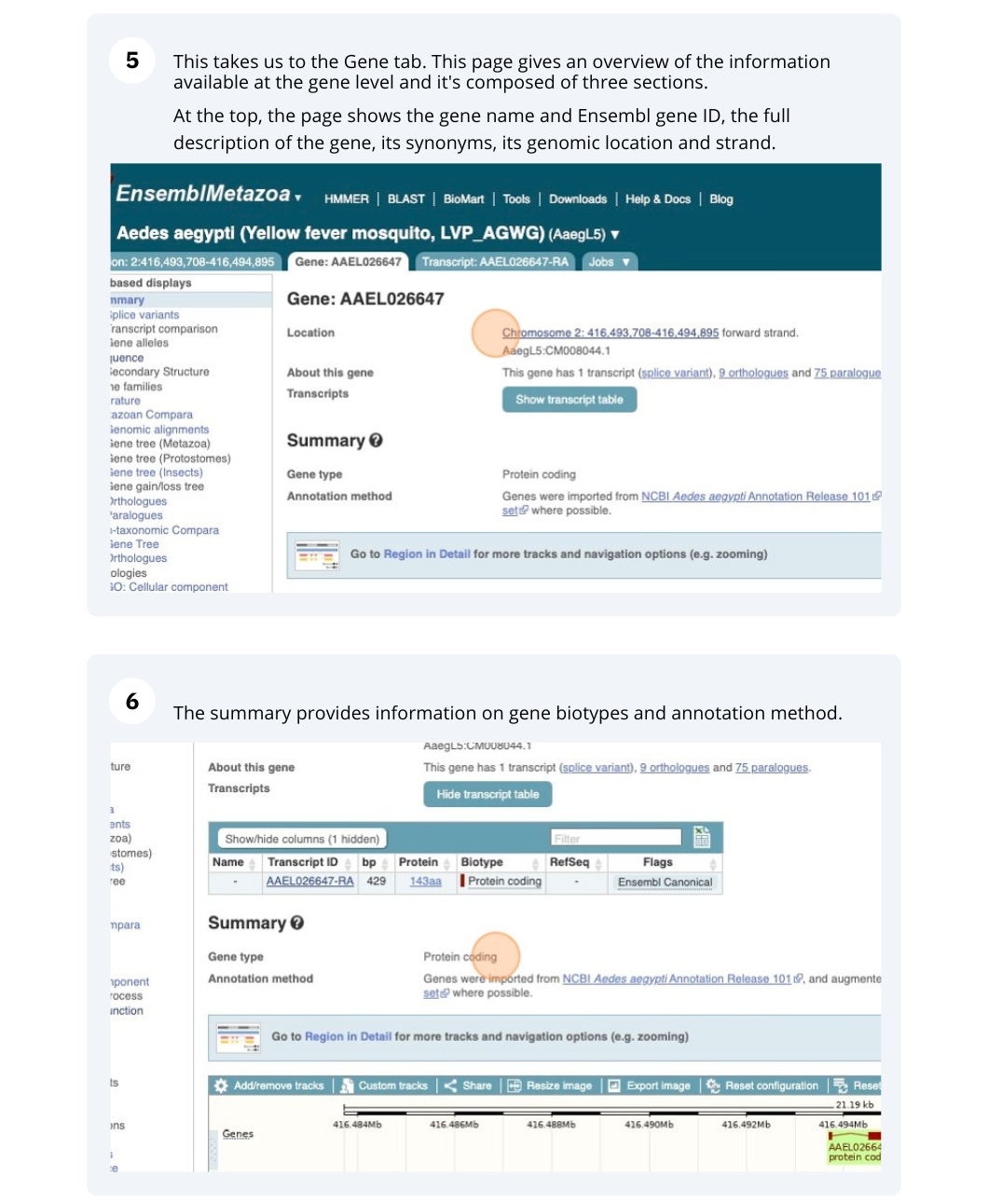
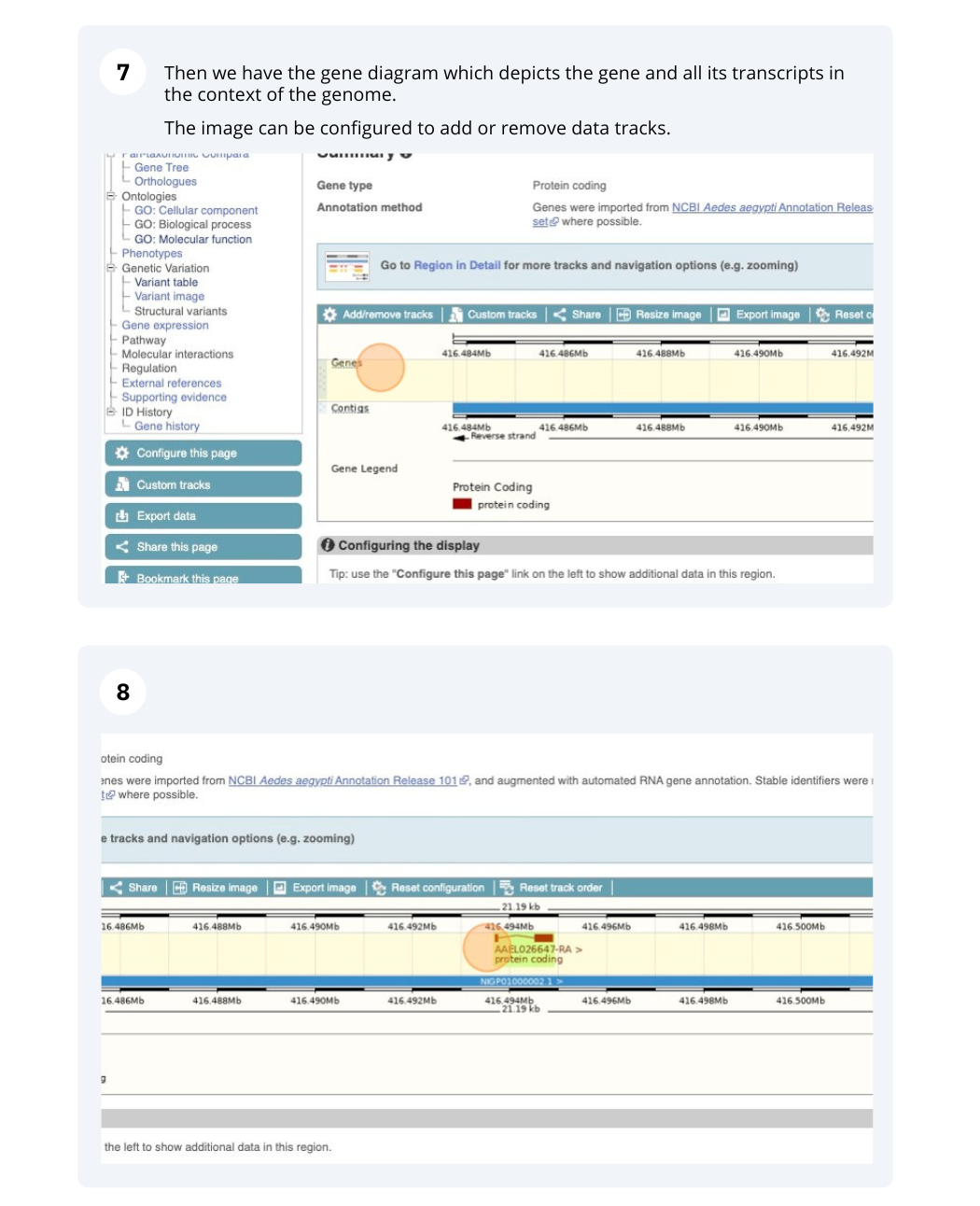
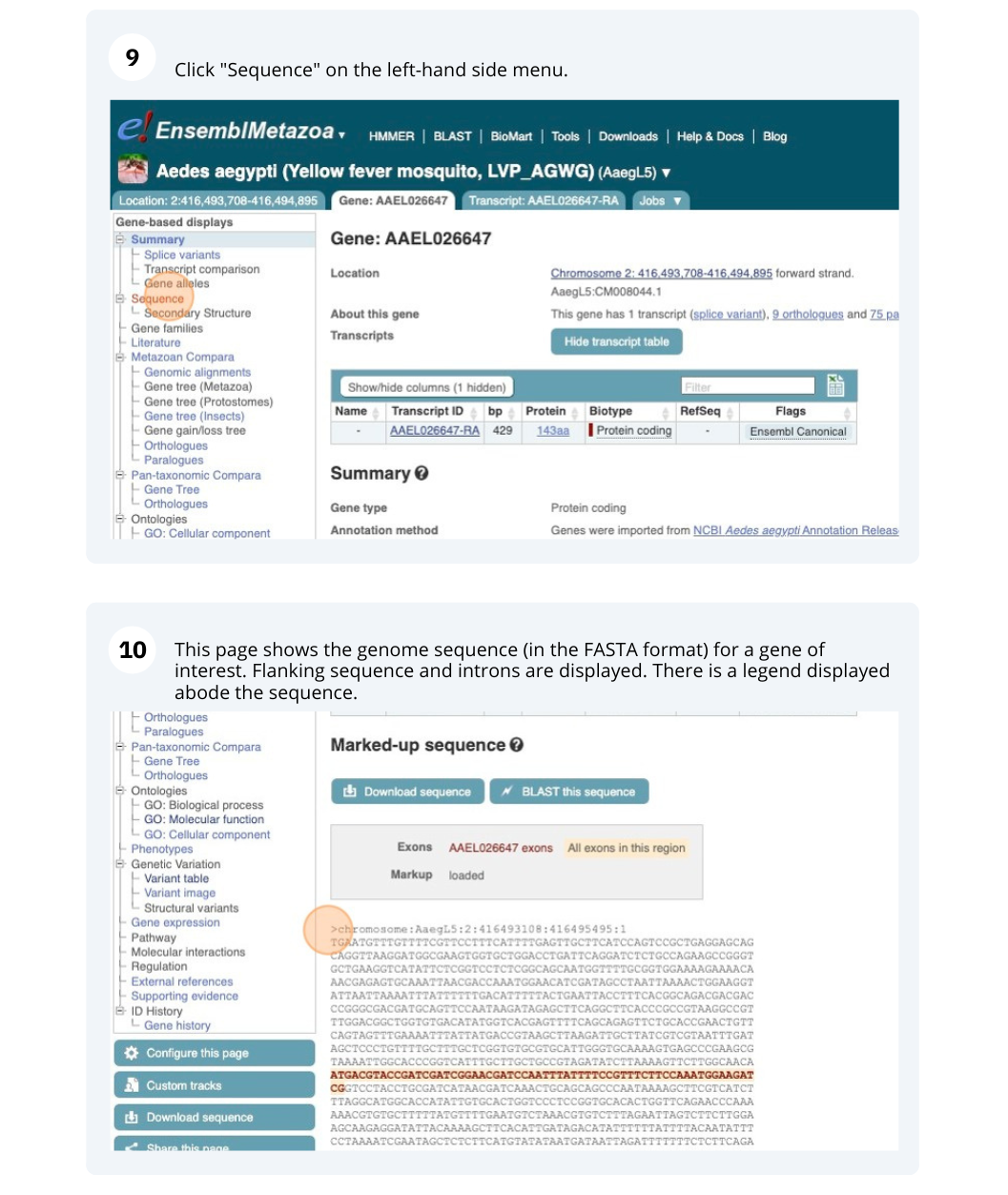
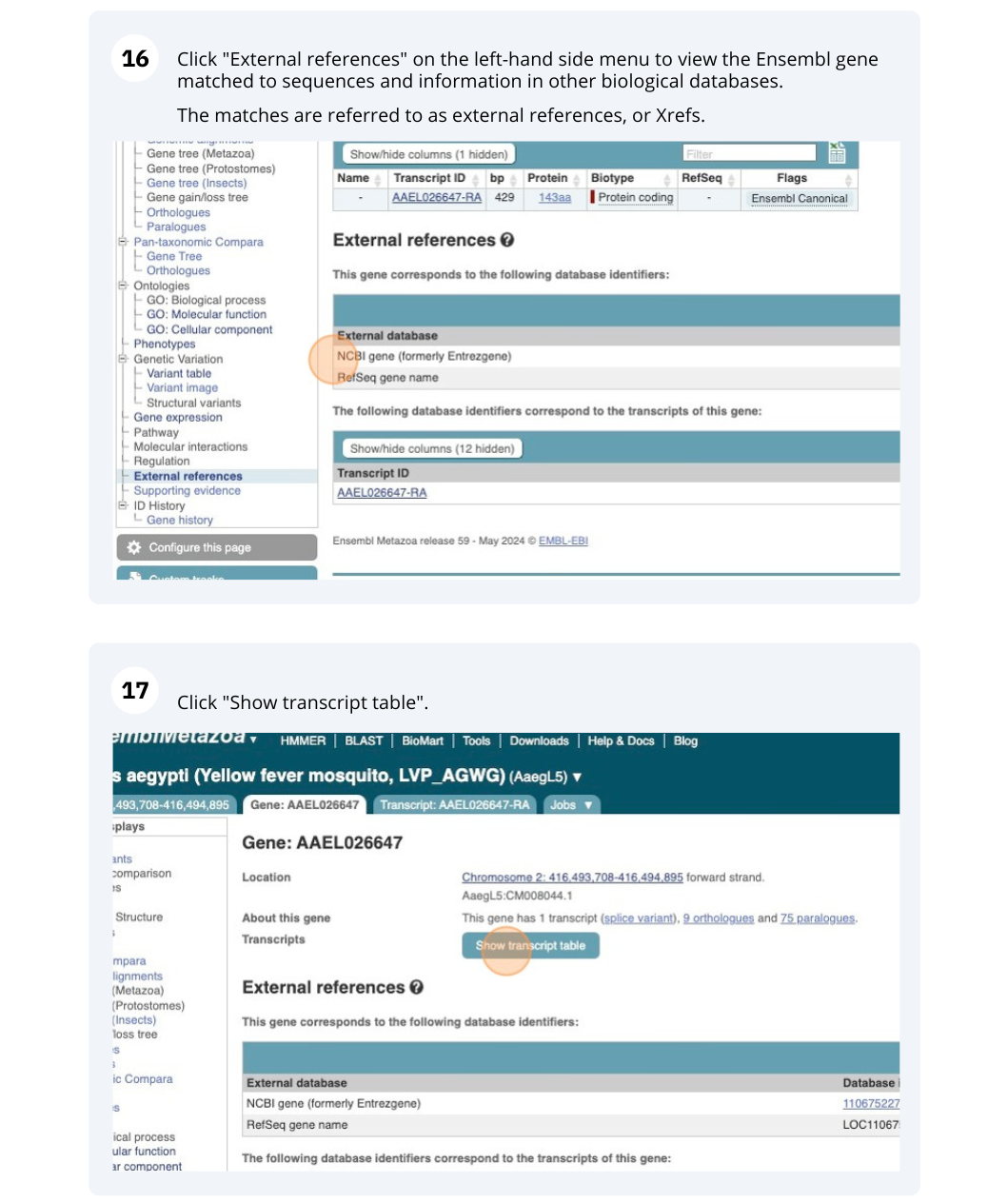
We’re going to look at the AAEL026647 gene in Aedes aegypti (Yellow fever mosquito, LVP_AGWG) to find out information about it and its transcript.
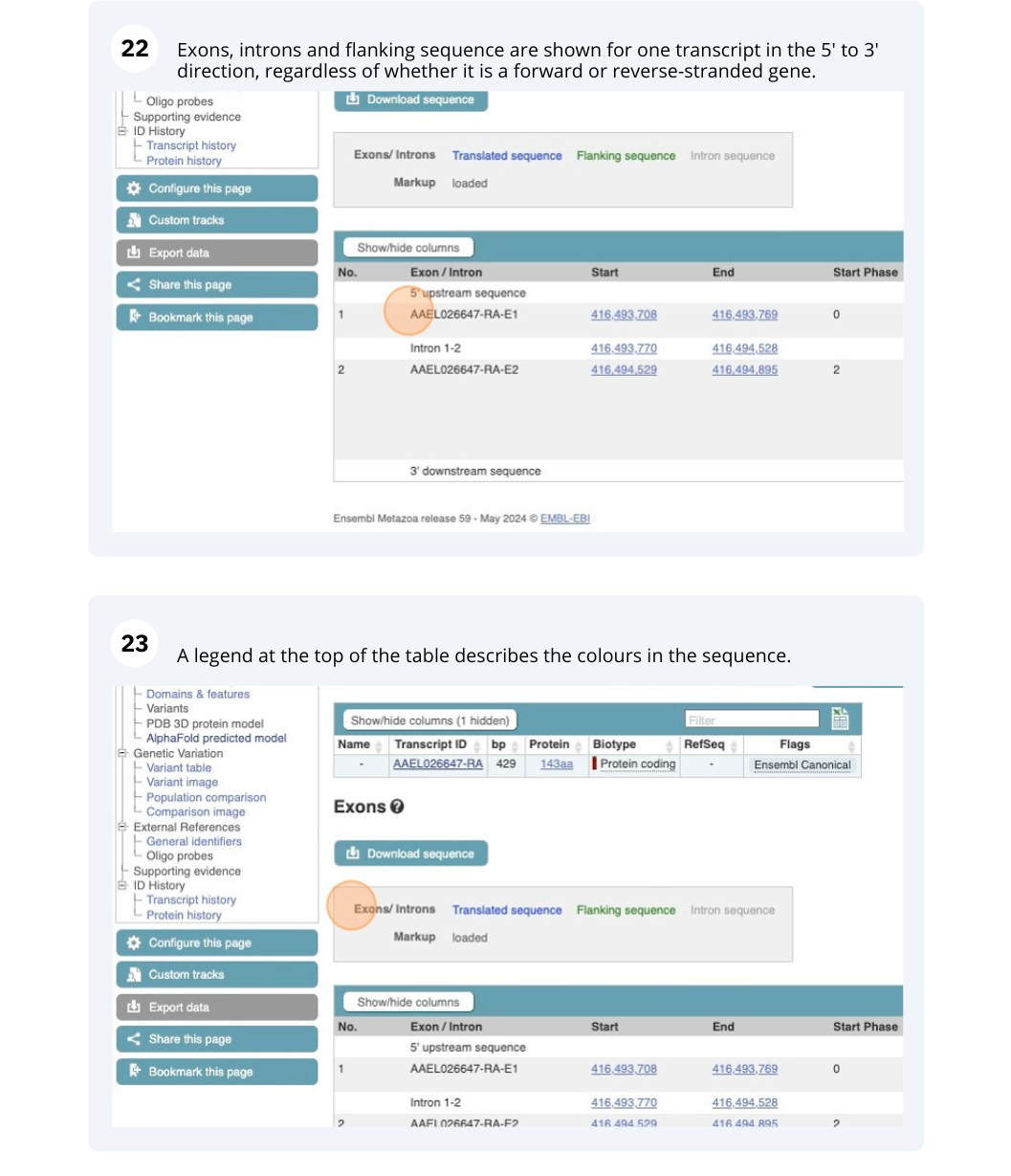
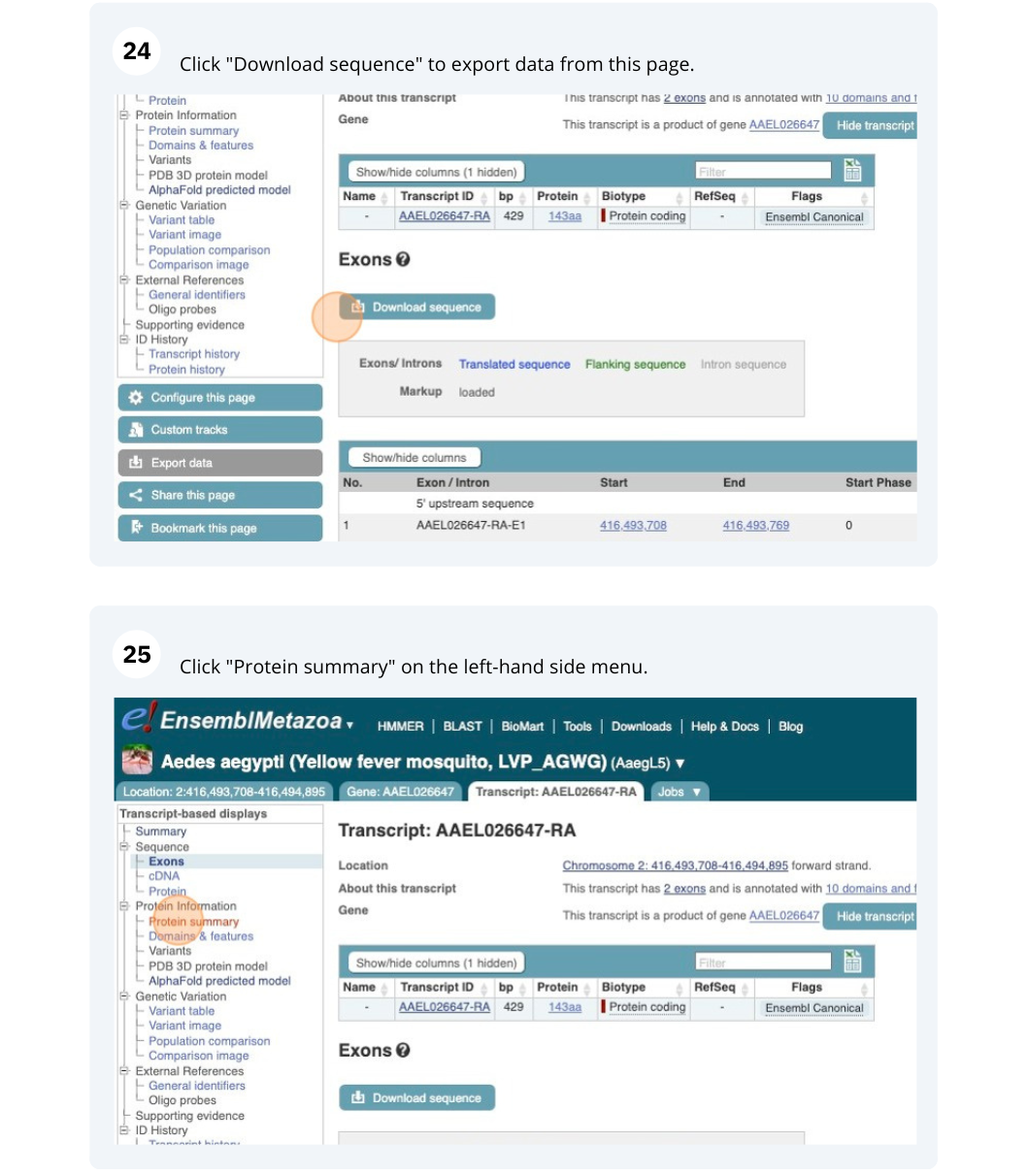
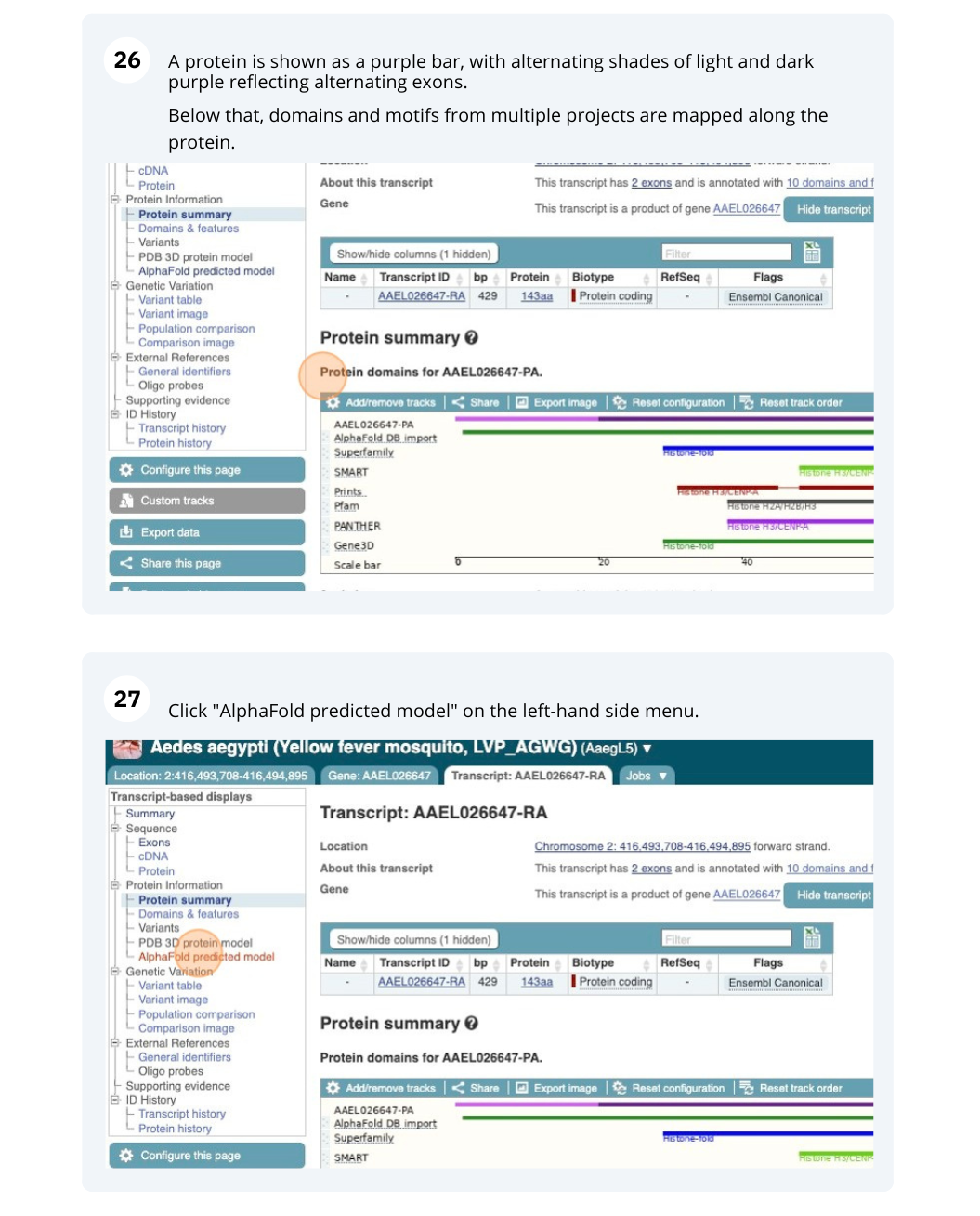
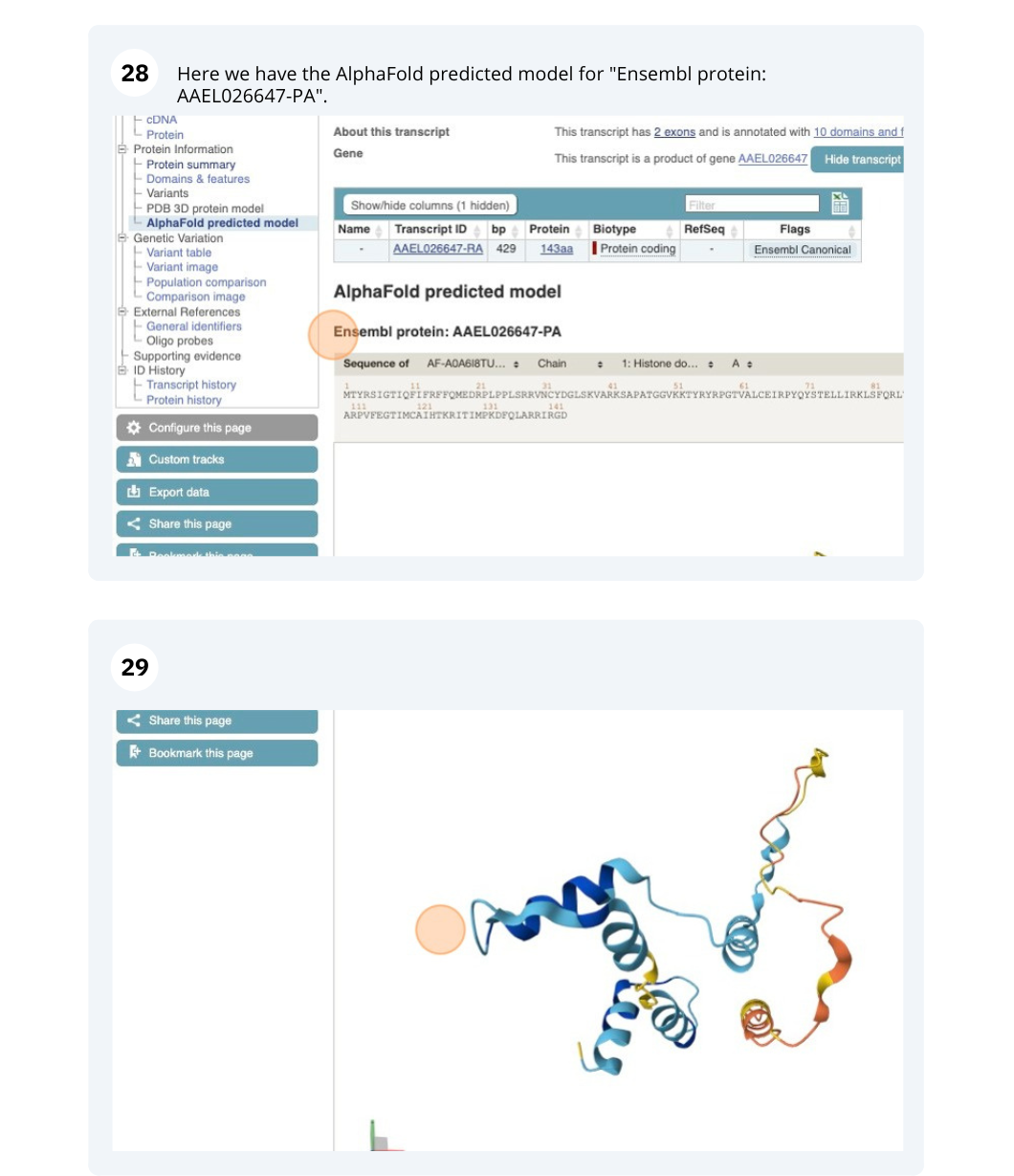
Exploring a leaf-cutter ant gene
-
Find the Atta cephalotes LOC105618535 gene on Ensembl Metazoa.
-
How long is its transcript? How long is the protein it encodes? How many exons does it have? Are any of the exons completely or partially untranslated?
-
Export the sequence of the gene, cDNA and protein in FASTA format.
-
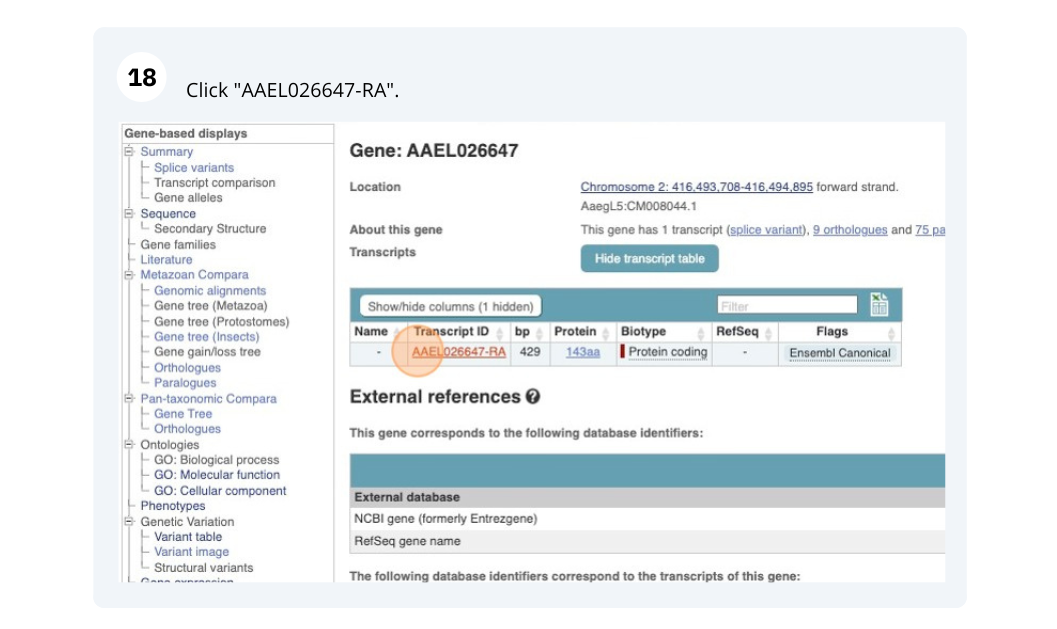
Go to the Ensembl Metazoa homepage. Select Atta cephalotes (Leaf-cutter ant) from the species list and type LOC105618535 in the search box. Click Go. Click on LOC10561855.
-
Click on Show transcript table.
The transcript is 3451 base pairs and the length of the encoded protein is 810 amino acids.
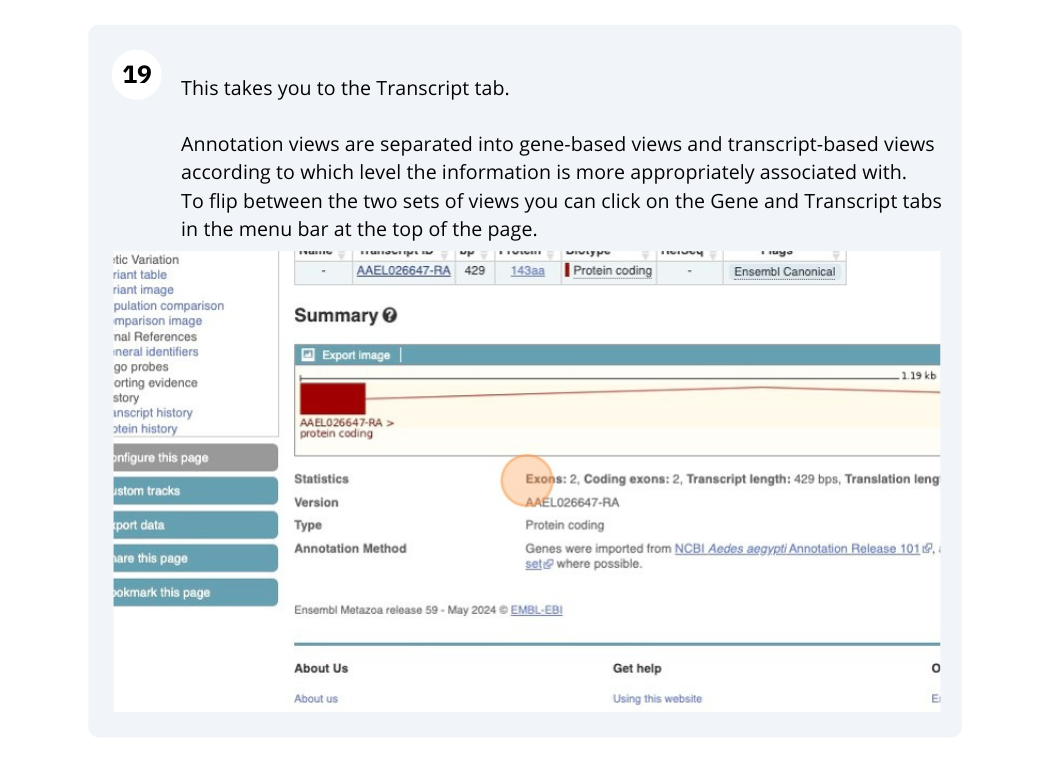
Click on the Ensembl Transcript ID XM_012200067.1 in the transcript table.
It has nine exons.
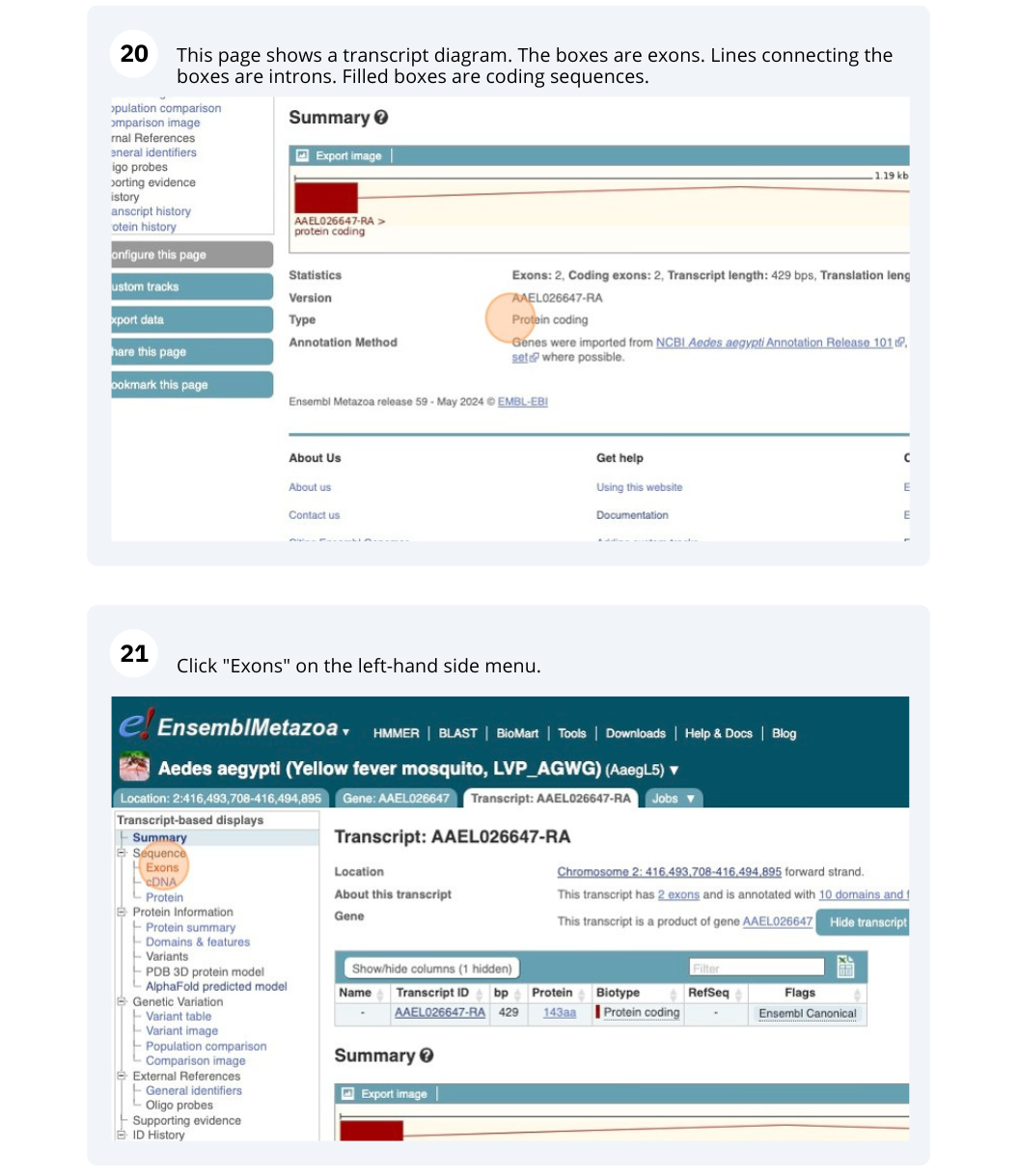
Click on Sequence - Exons in the side menu.
The last exon is partially untranslated (sequence shown in orange). This can also been seen from the fact that in the transcript diagram on the Gene summary and Transcript summary pages the boxes representing the last exon is partially unfilled.
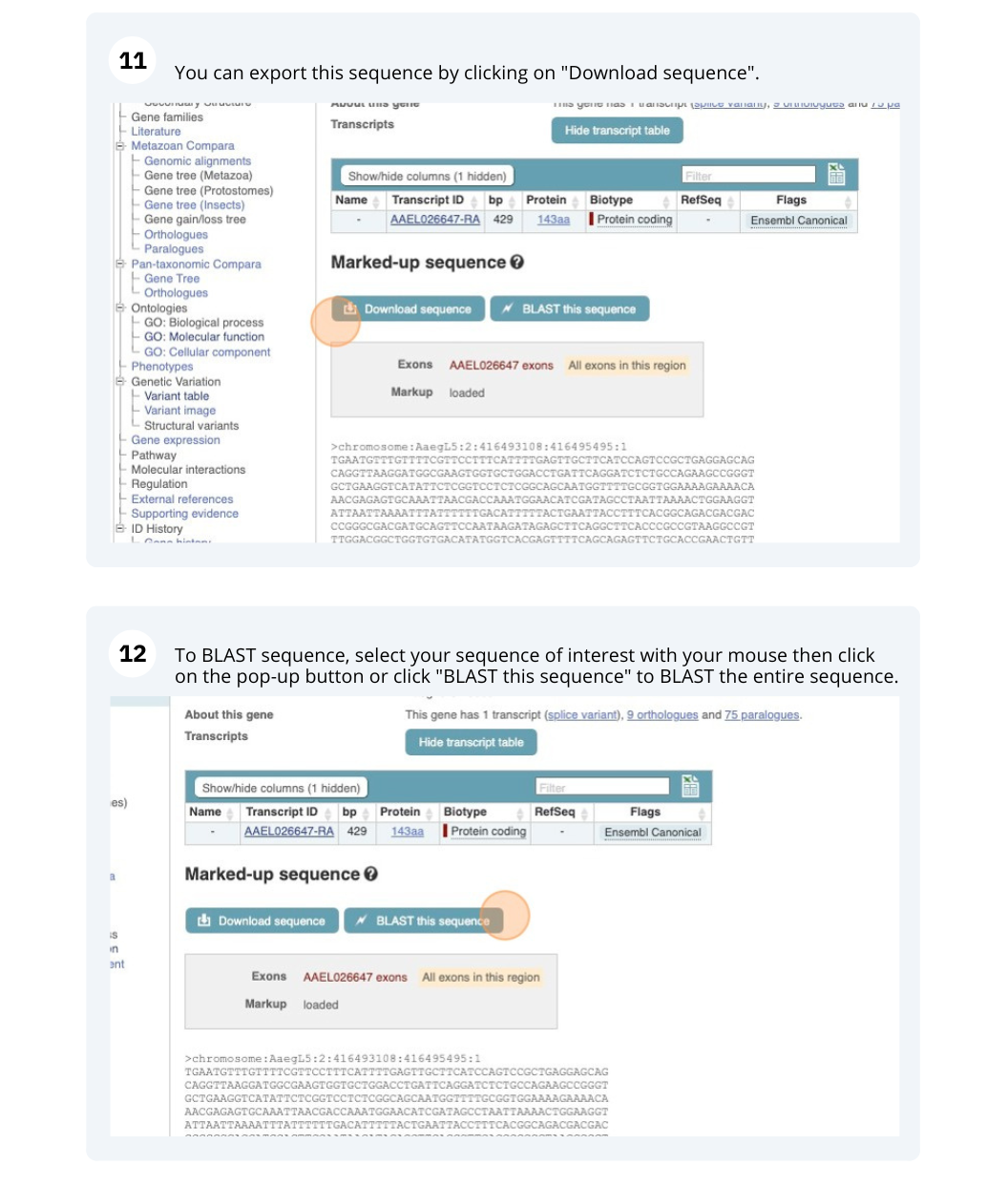
- Click on the blue Export data button. Under Options for FASTA sequence, select Genomic: Unmasked, cDNA and Peptide sequence. Click Next>. Click on Text.
This returns three sequences (one gene, one transcript and one protein sequence).
Exploring an Anopheles gambiae gene
Start in metazoa.ensembl.org/index.html and select the Anopheles gambiae (African malaria mosquito, PEST) genome.
(a) What GO: biological process terms are associated with the para gene?
(b) How many protein coding transcripts does this gene have? View all of these in the transcript comparison view.
(c) Go to the transcript tab for the transcript, AGAP004707-RH. How many exons does it have? Which one is the longest?
(a) Go to metazoa.ensembl.org/index.html. Click on Anopheles gambiae (African malaria mosquito, PEST) from the popular species list.
Search for para and click on the AGAP004707 link in the results.
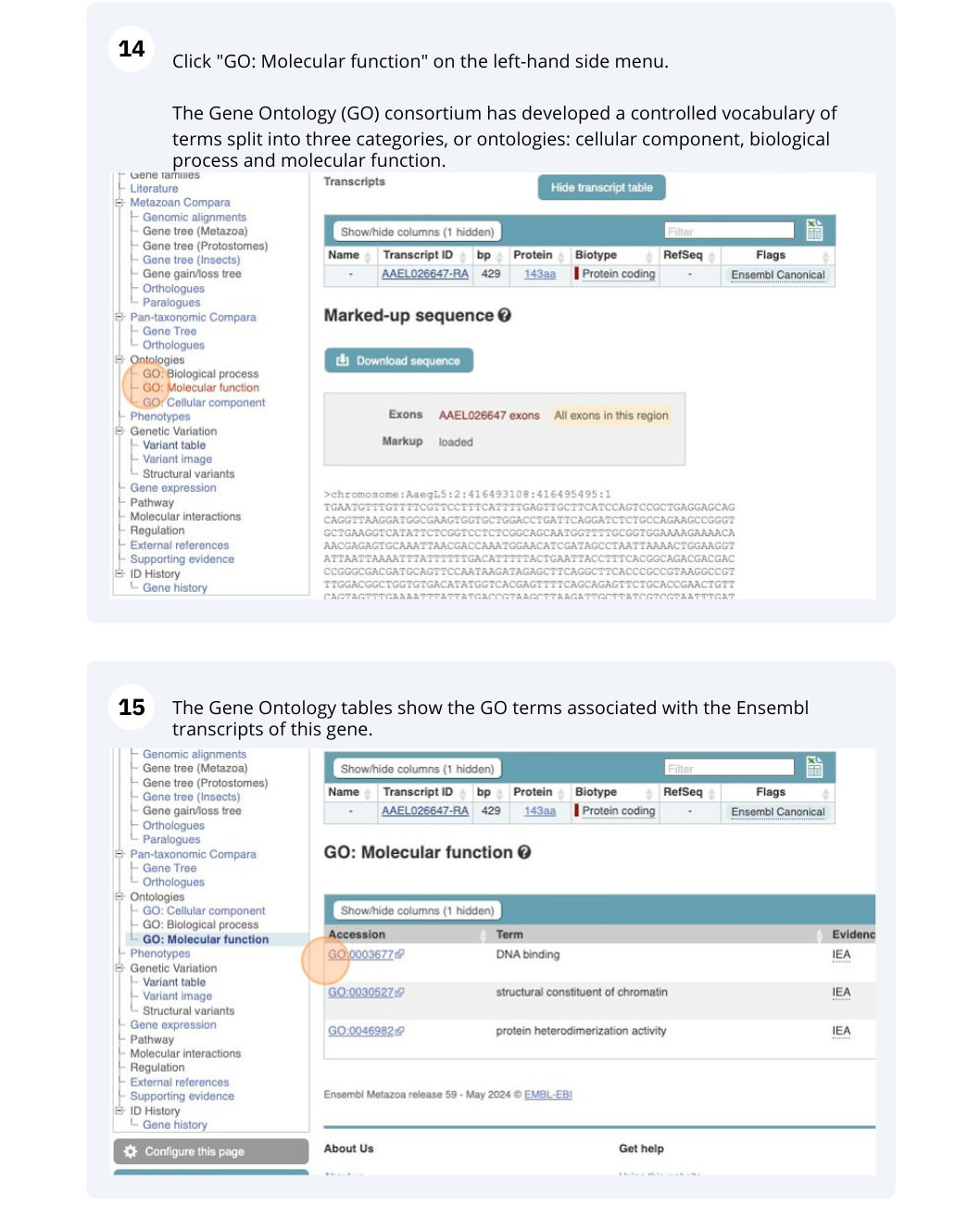
Click on GO: biological process in the side menu.
There are nine GO terms listed. GO:0006811 ion transport, GO:0006814 sodium ion transport and GO:0019228 neuronal action potential are some of the terms listed.
(b) If the transcript table is hidden, click on Show transcript table to see it.
There are 13 protein coding transcripts.
Click on Transcript comparison in the left hand menu. Click on Select transcripts. Either select all the transcripts labelled protein coding one-by-one, or click on the drop down and select Protein coding. Close the menu.
(c) Click on the transcript named AGAP004707-RH. Click on Exons in the left hand menu.
There are 32 exons, of which exon 32 is longest with 1,017 bp.
Exploring a gene in Plasmodium falciparum
-
Find the Plasmodium falciparum 3D7 PF3D7_1145400 gene on Ensembl Protists. On which strand is this gene located? What are the coordinates of the gene?
-
How long is its transcript (in bp)? How long is the protein it encodes? How many exons does it have?
-
What is the Uniprot ID that maps to the translation of this transcript?
-
What are the GO:Biological process(es) associated with PF3D7_1145400?
- Go to the Ensembl Protists homepage. Select Plasmodium falciparum from the species list and type
PF3D7_1145400in the search box. Click Go. Click on PF3D7_1145400 in the search results. You can find the strand orientation and coordinates in the gene Summary page.PF3D7_1145400 is located on the reverse strand of chromosome 11 between 1,800,544 and 1,803,550.
- Click on Show transcript table.
The transcript is 2,514 base pairs and the length of the encoded protein is 837 amino acids.
Click on the transcript ID CZT99117 in the transcript table.
It has four exons.
- You can find this information in a number of places: the transcript table, External references on the Gene tab or General identifiers on the Transcript tab.
The UniProt ID that maps to protein encoded by the PF3D7_1145400 transcript is Q8IHR4.
- Click on GO: Biological process in the side menu of the Gene tab.
The PF3D7_1145400 gene is involved in mitochondrial fission.
Variation
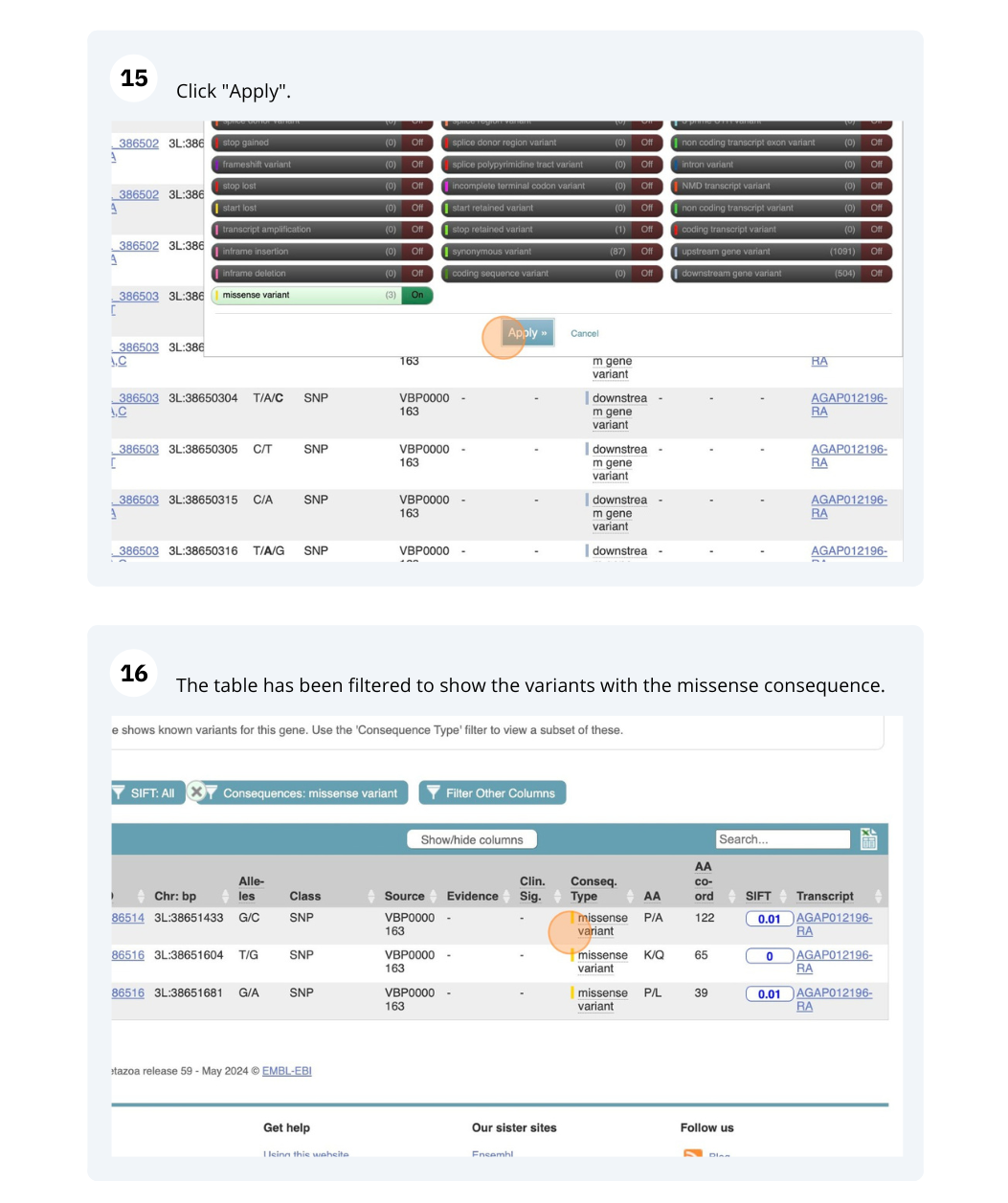
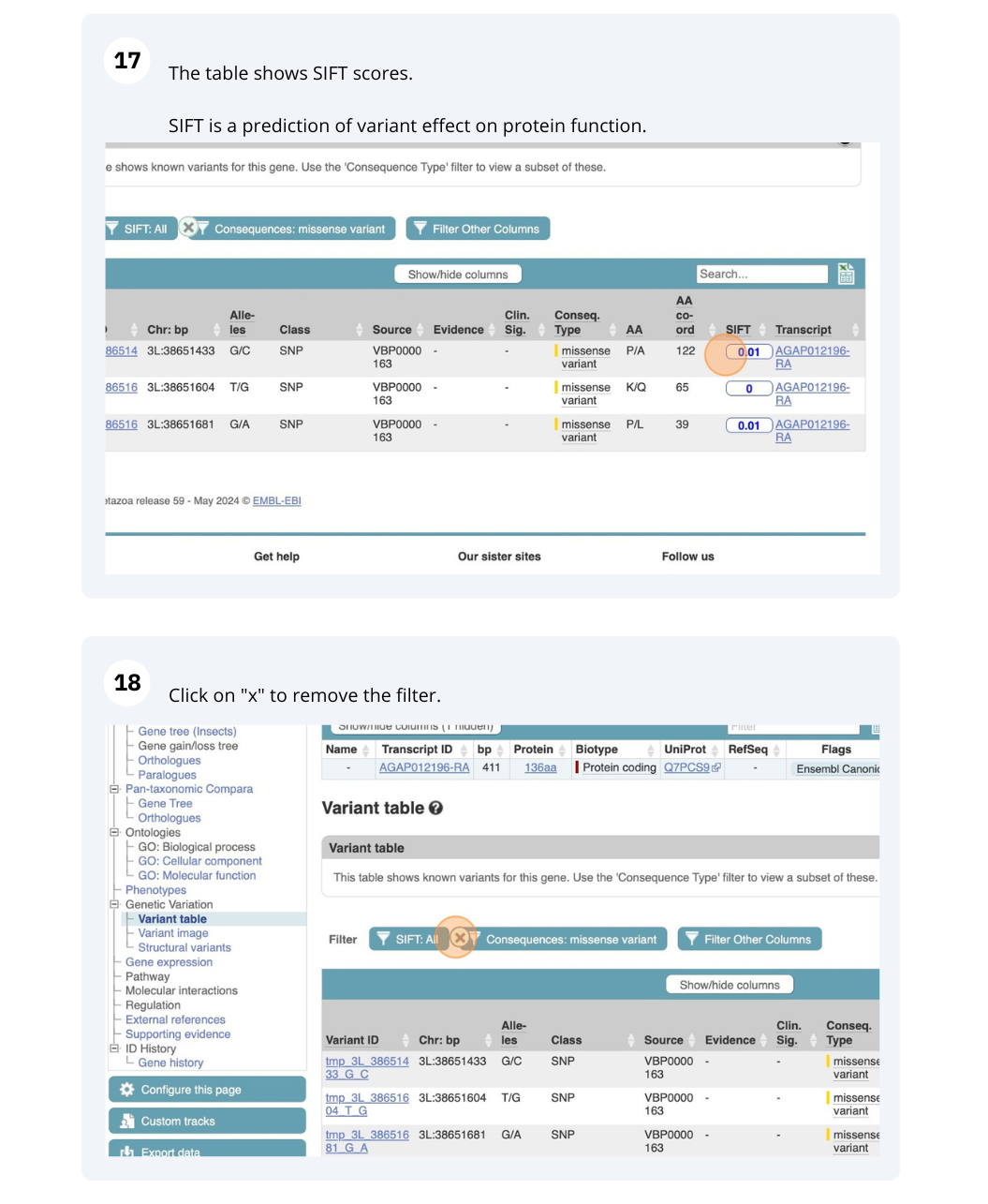
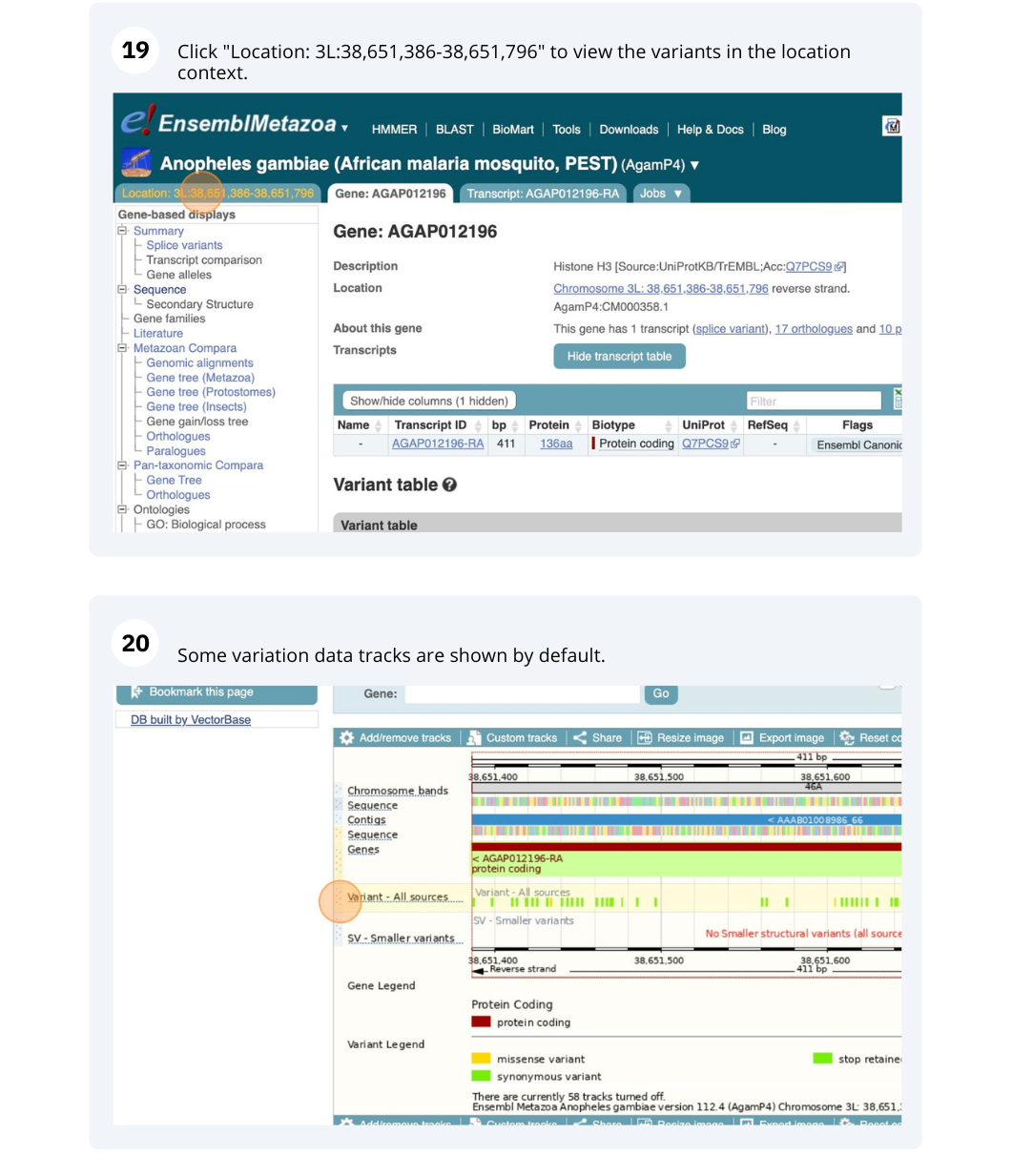
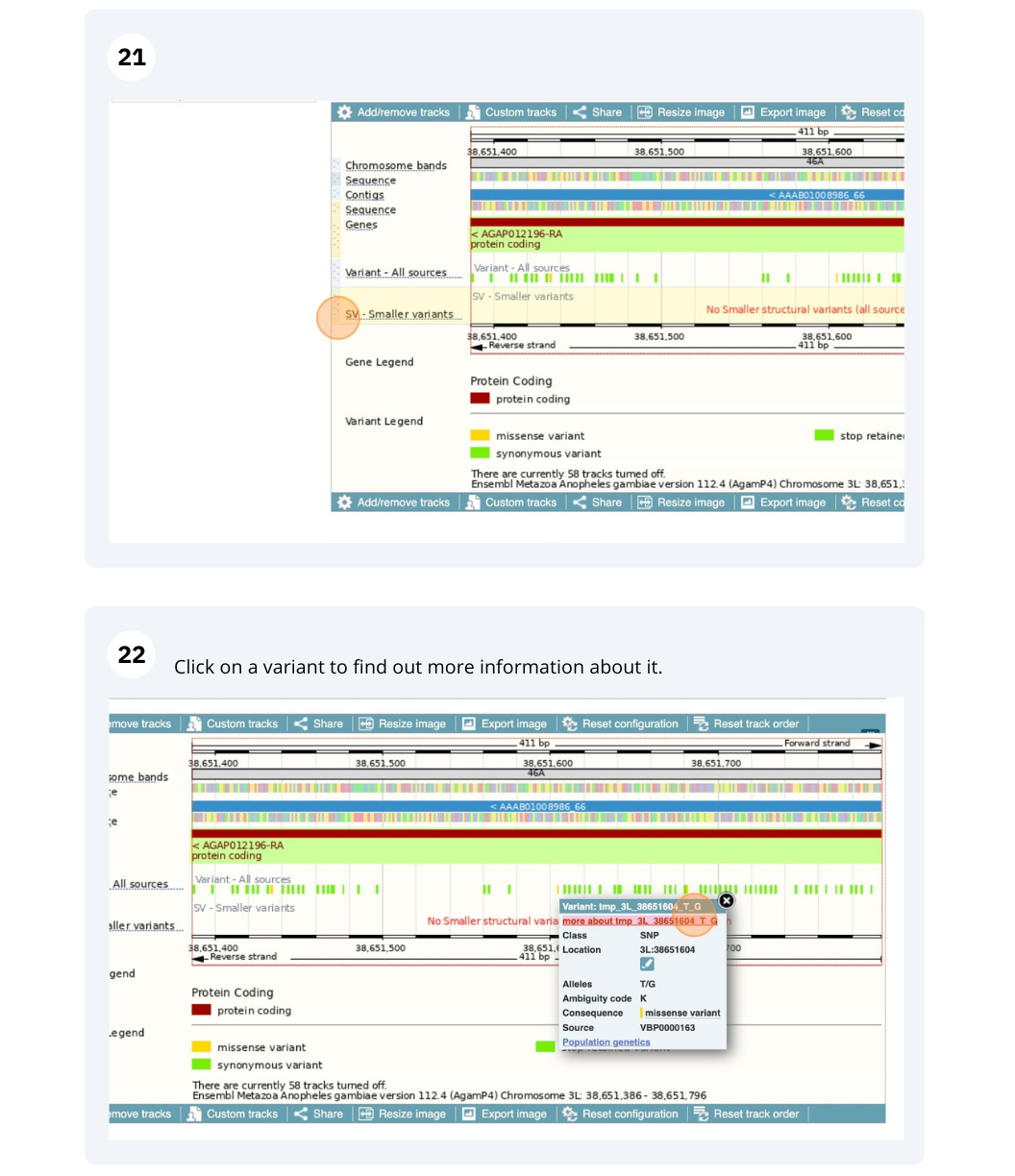
We are going to look at a gene AGAP012196 in Anopheles gambiae (African malaria mosquito, PEST)to find variants in the gene.
We will look at the region of AGAP012196 to find variants in the region.
We will look at a variant tmp_3L_38651604_T_G to find more information about it.
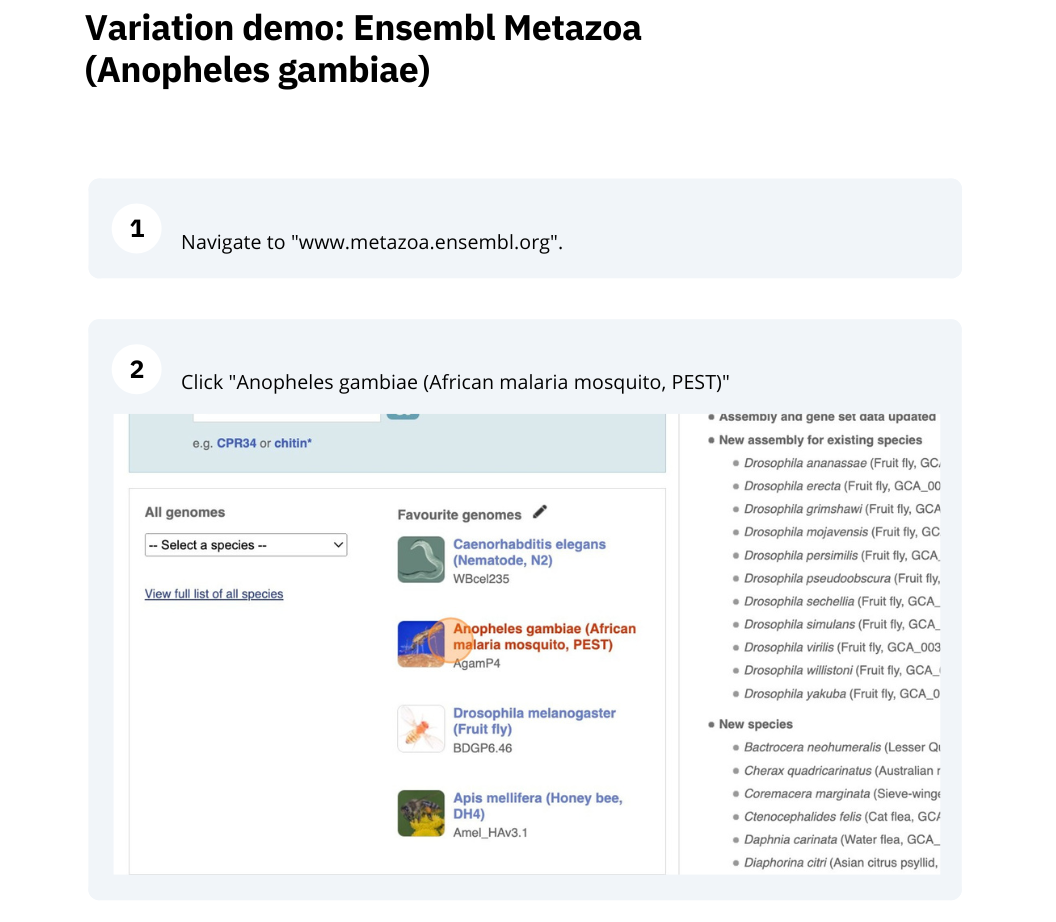
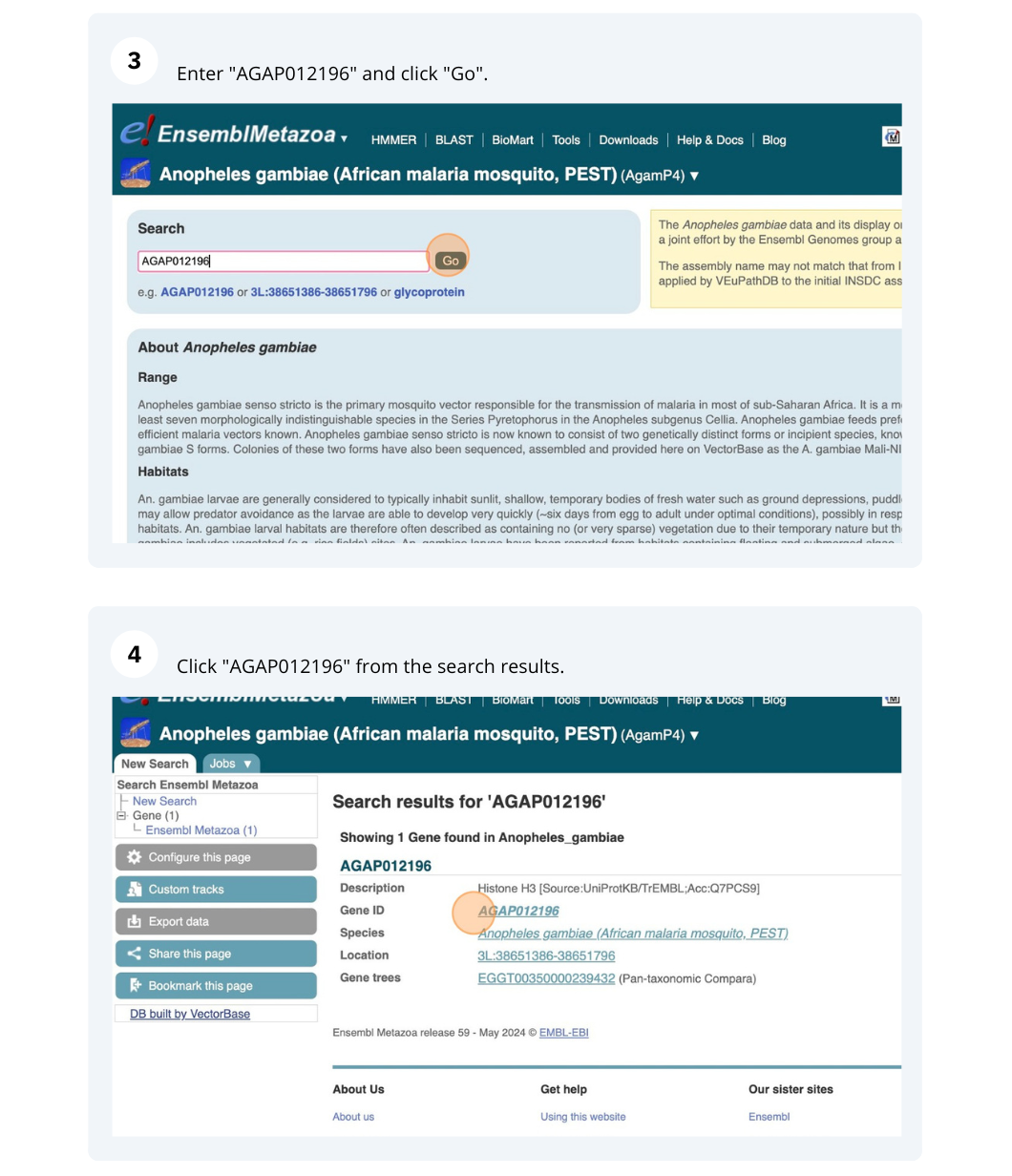
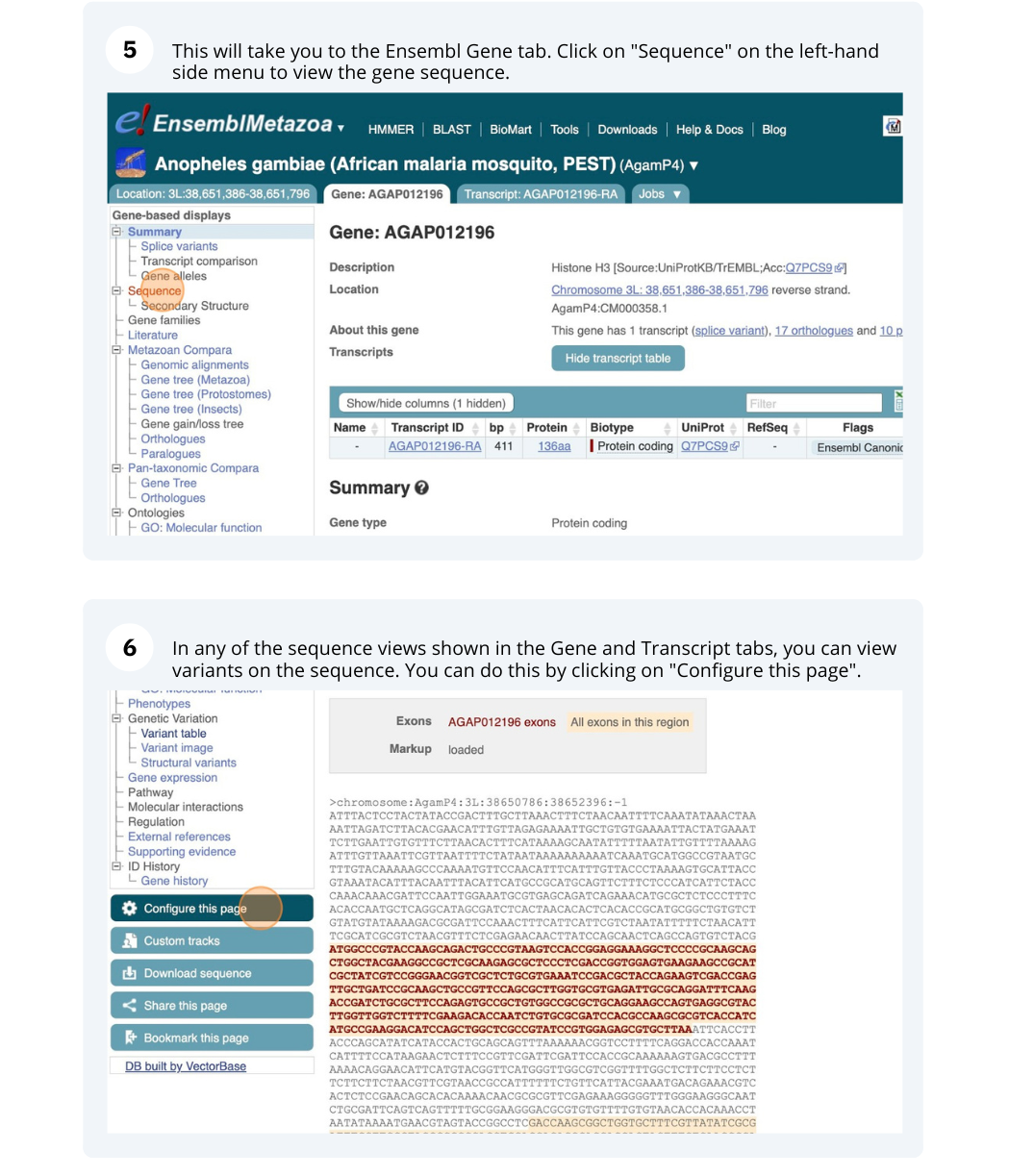
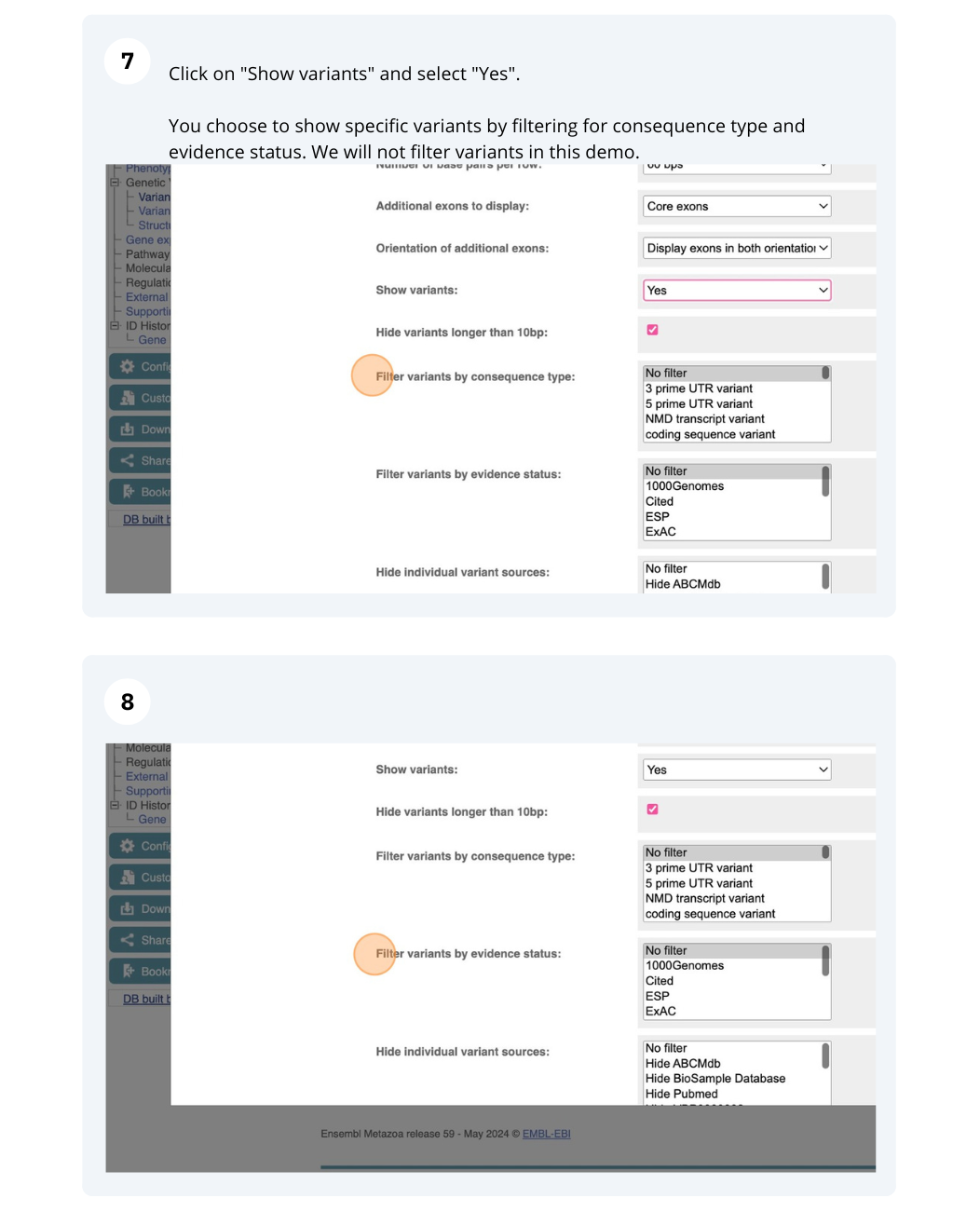
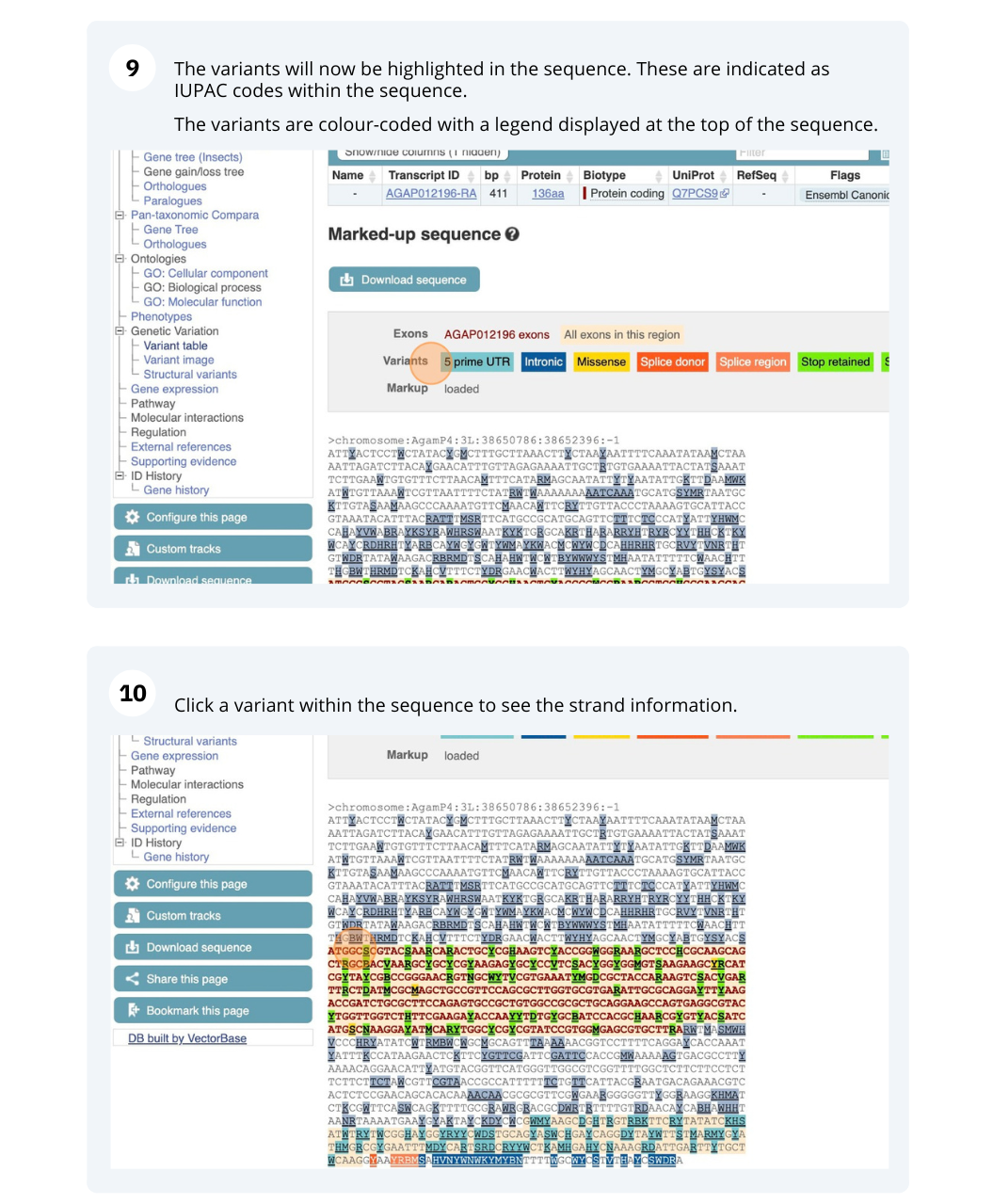
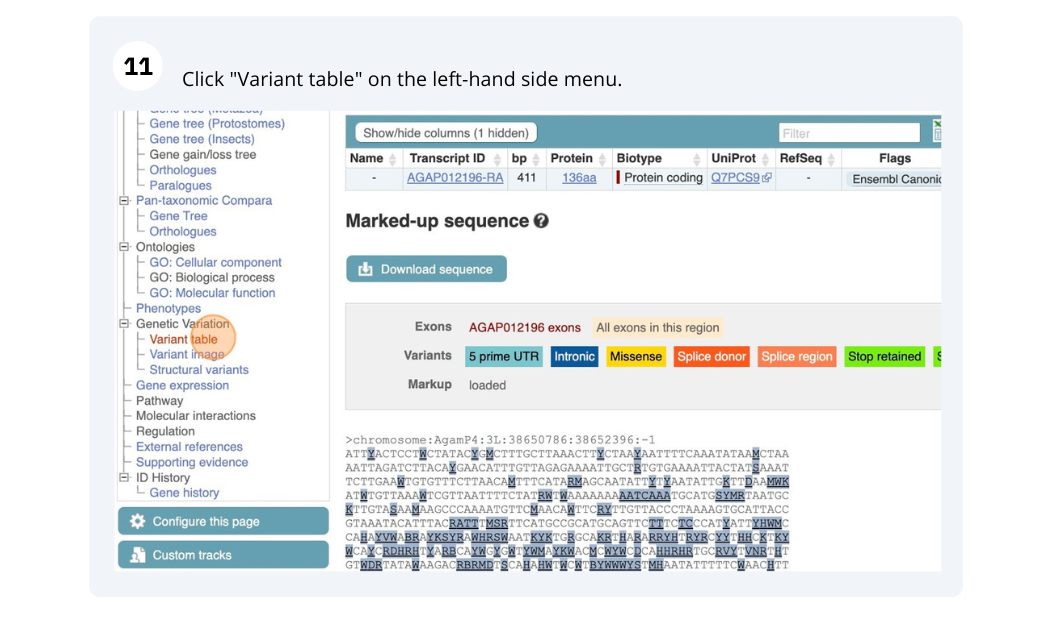
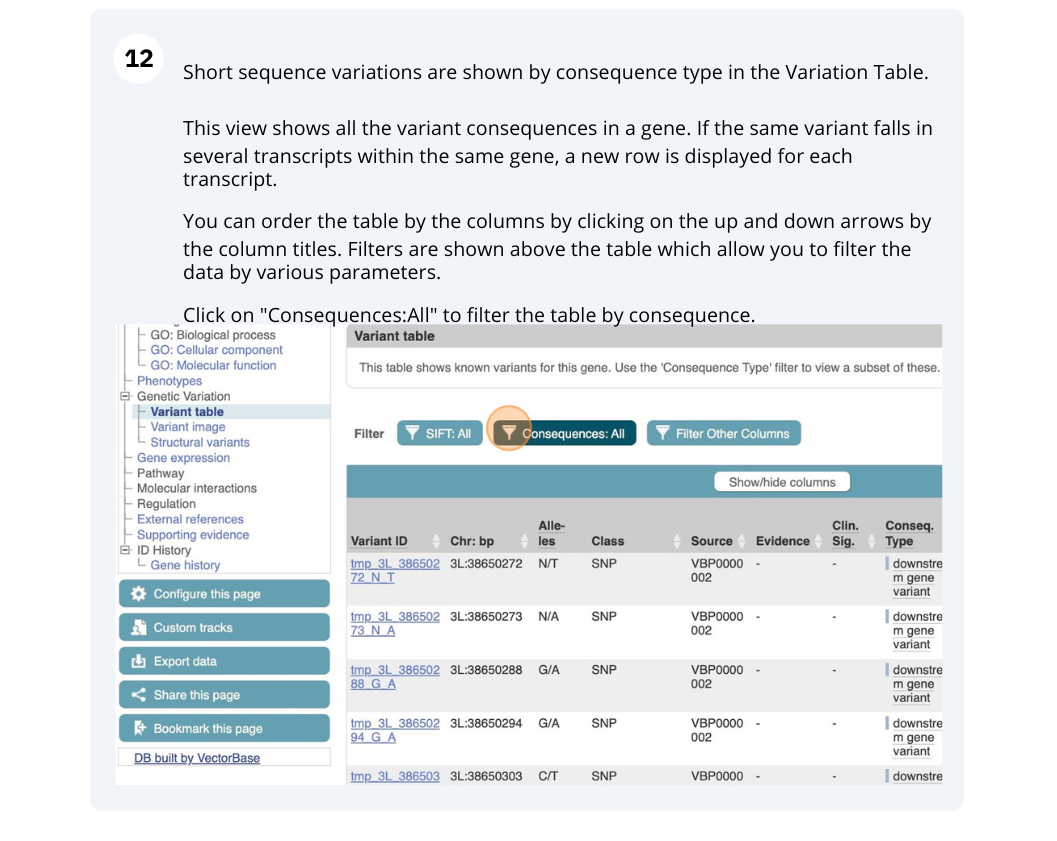
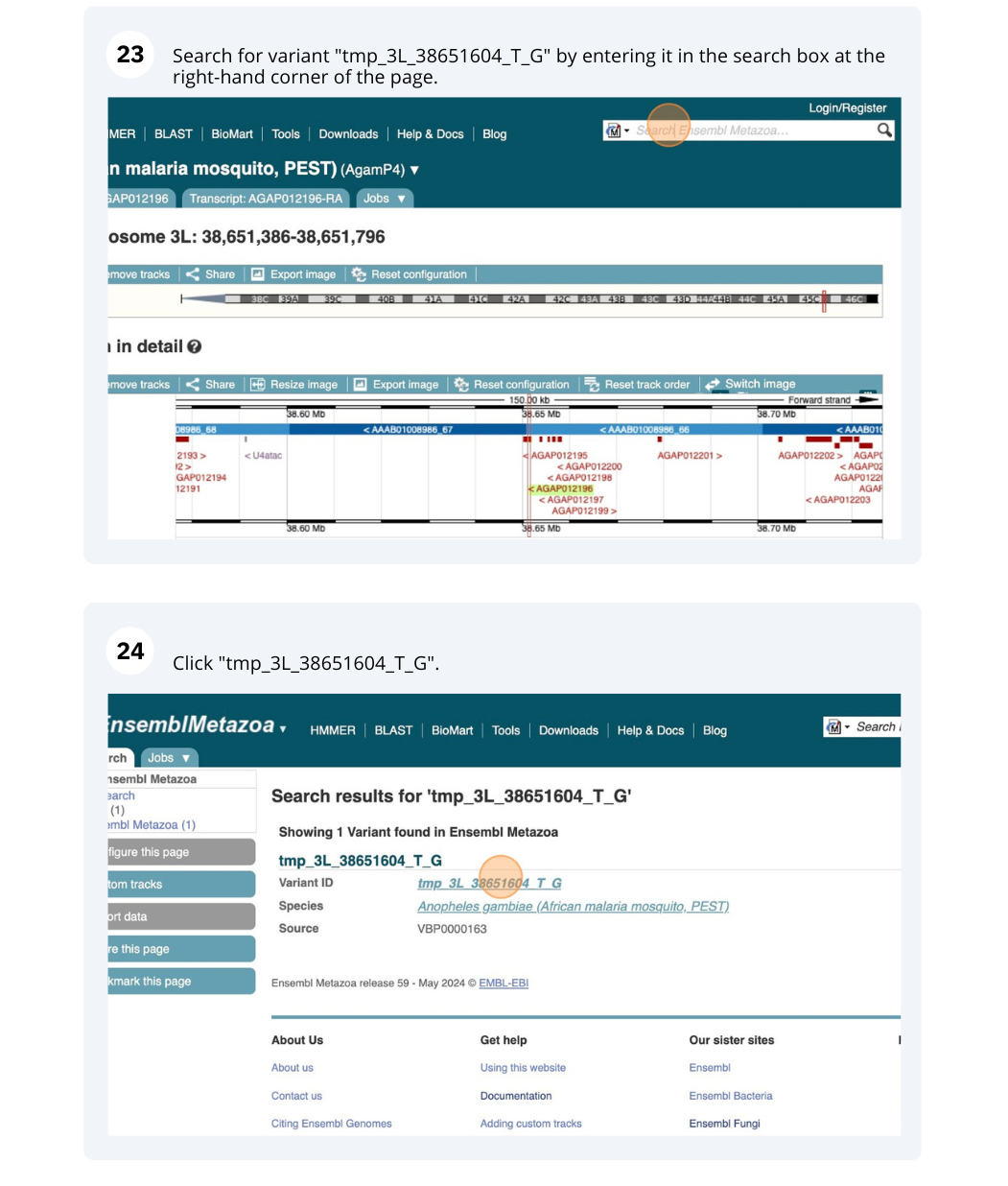
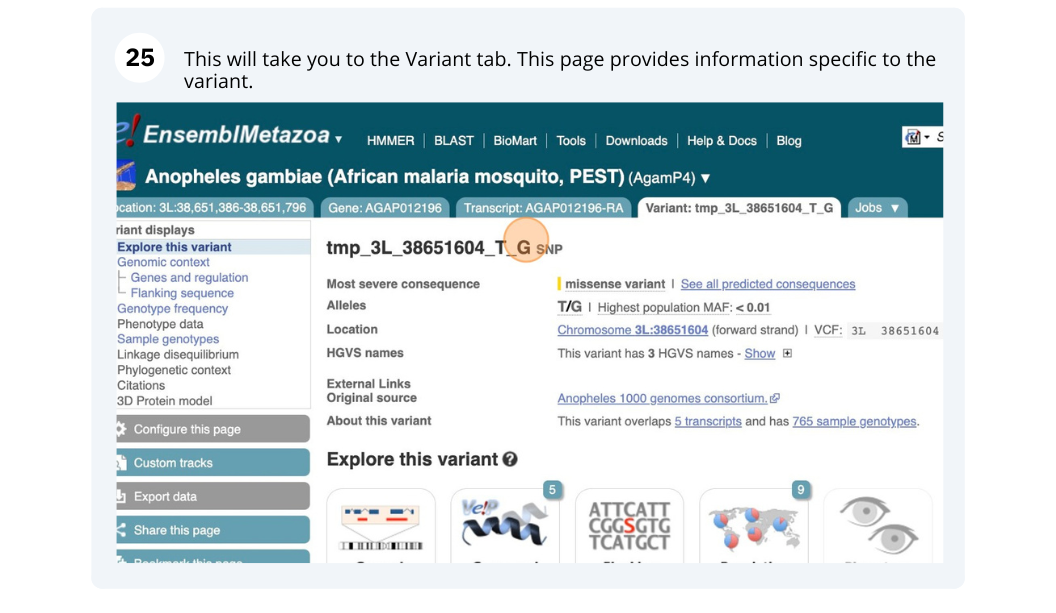
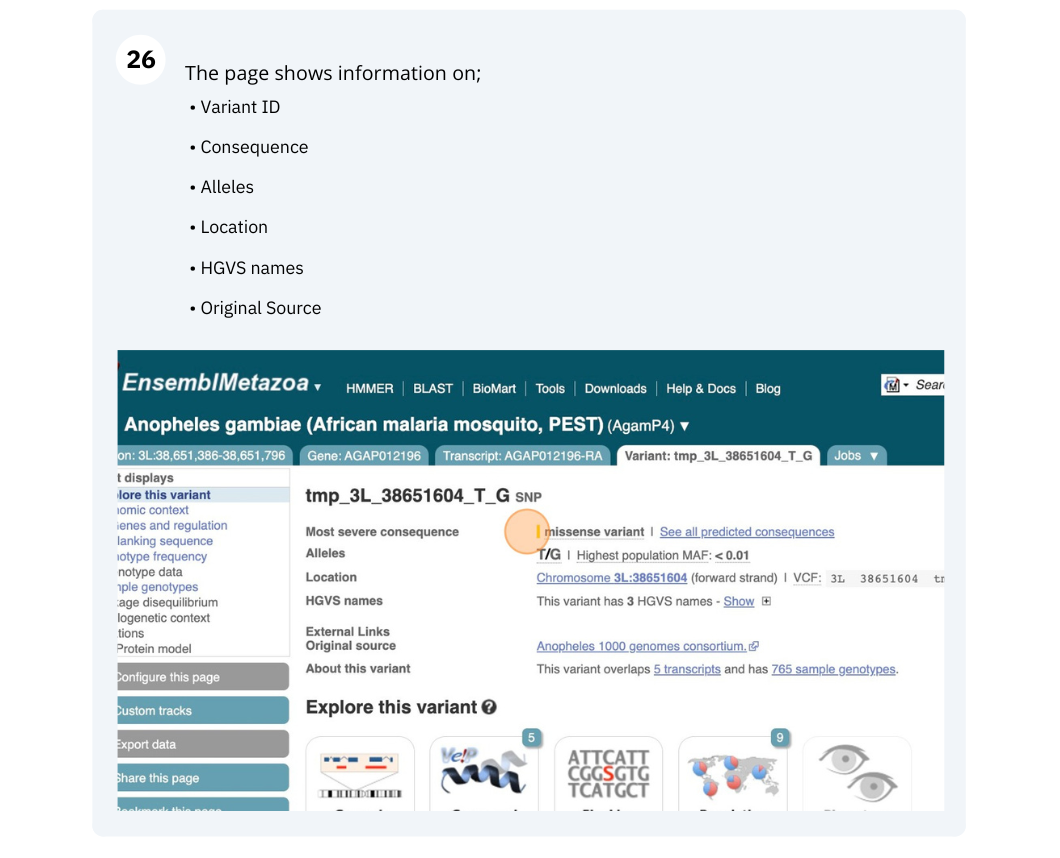
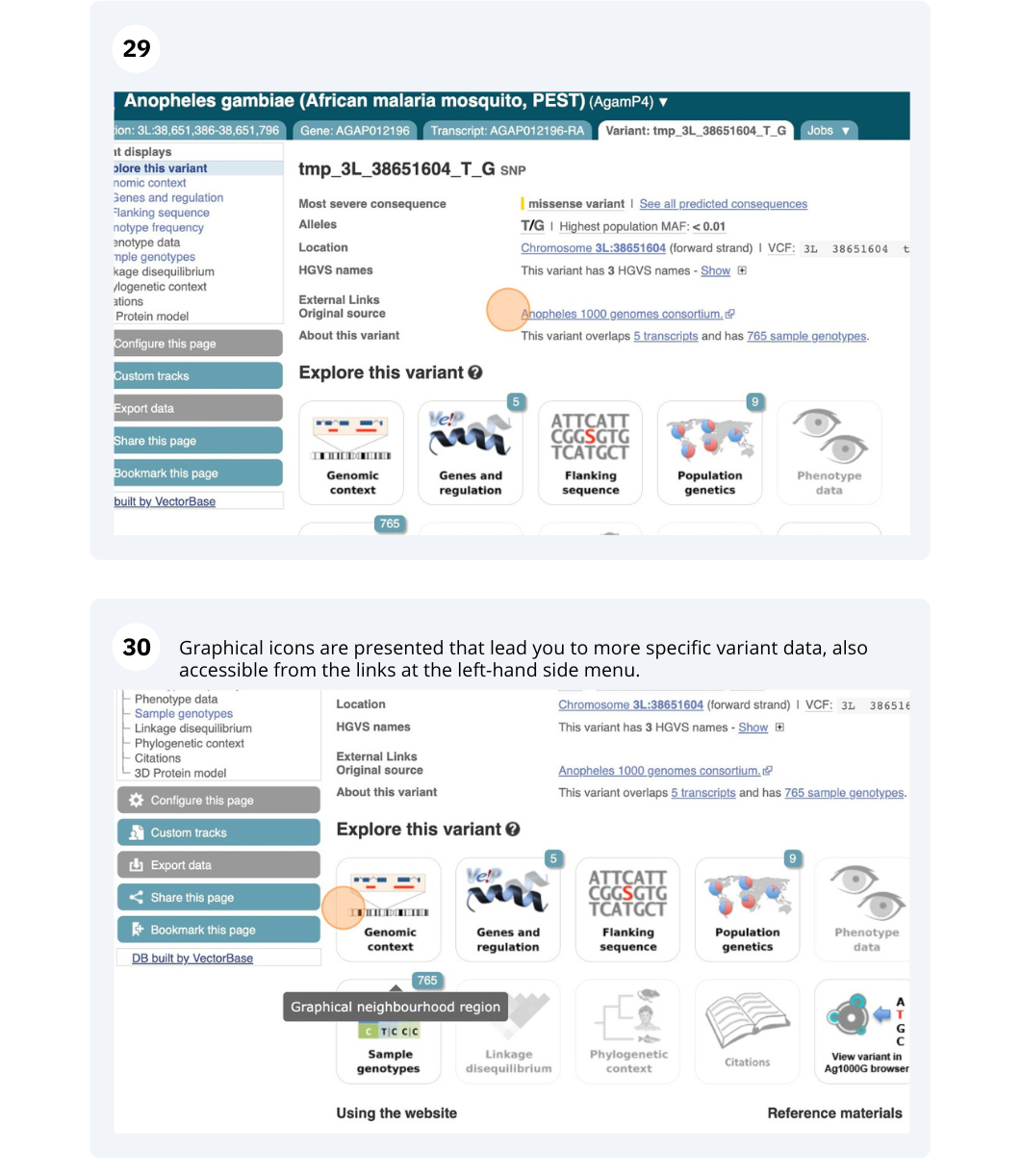
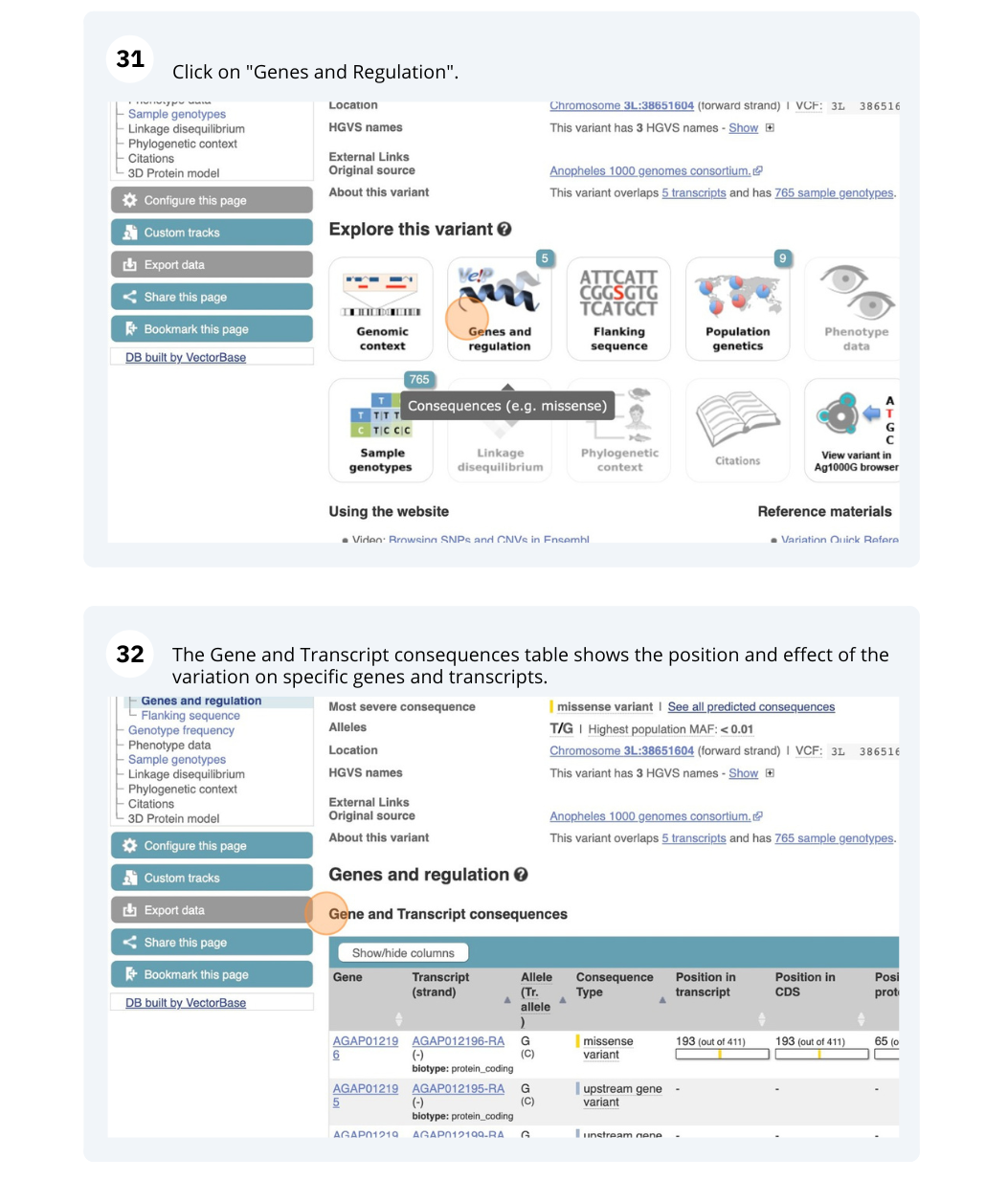
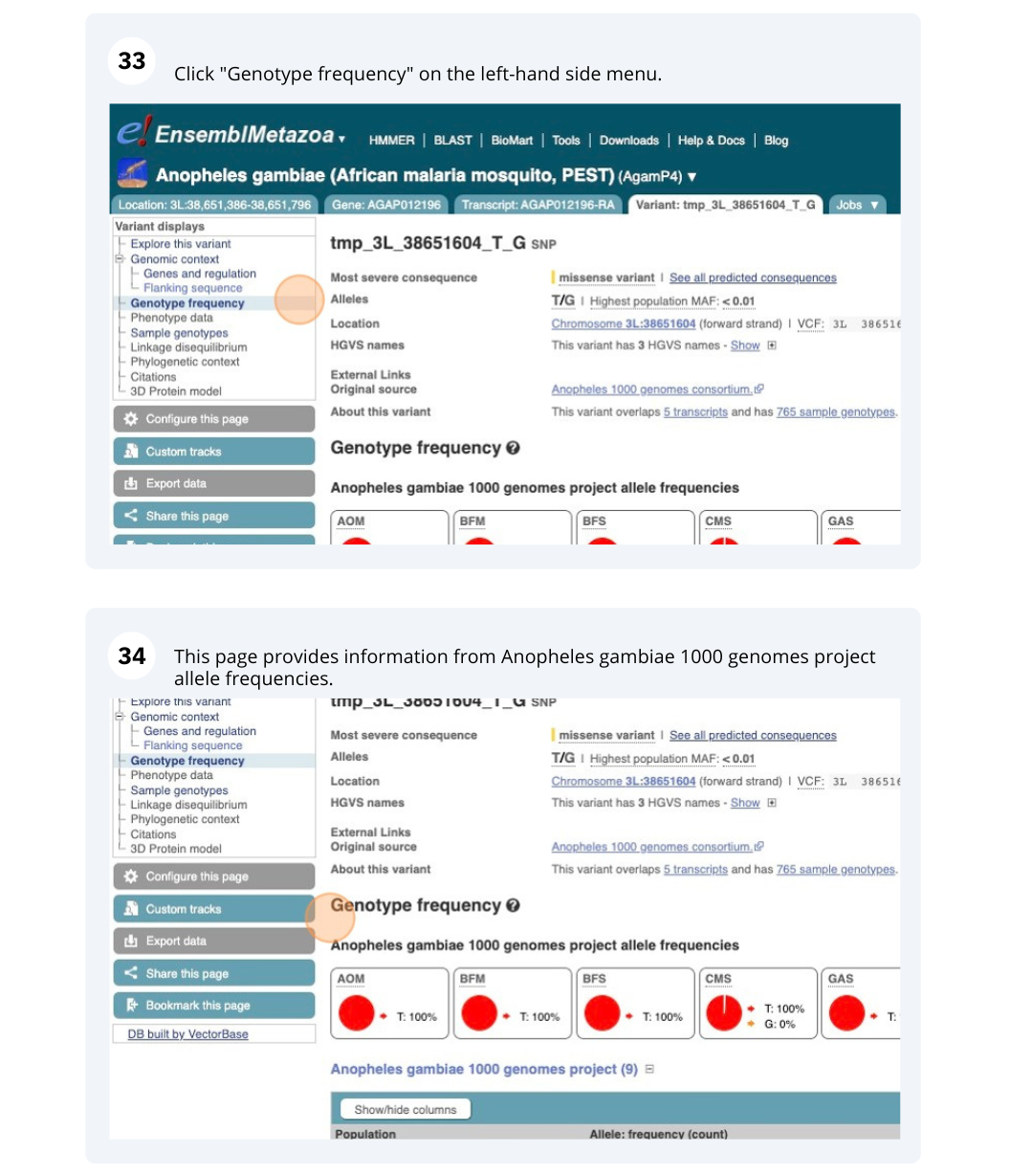
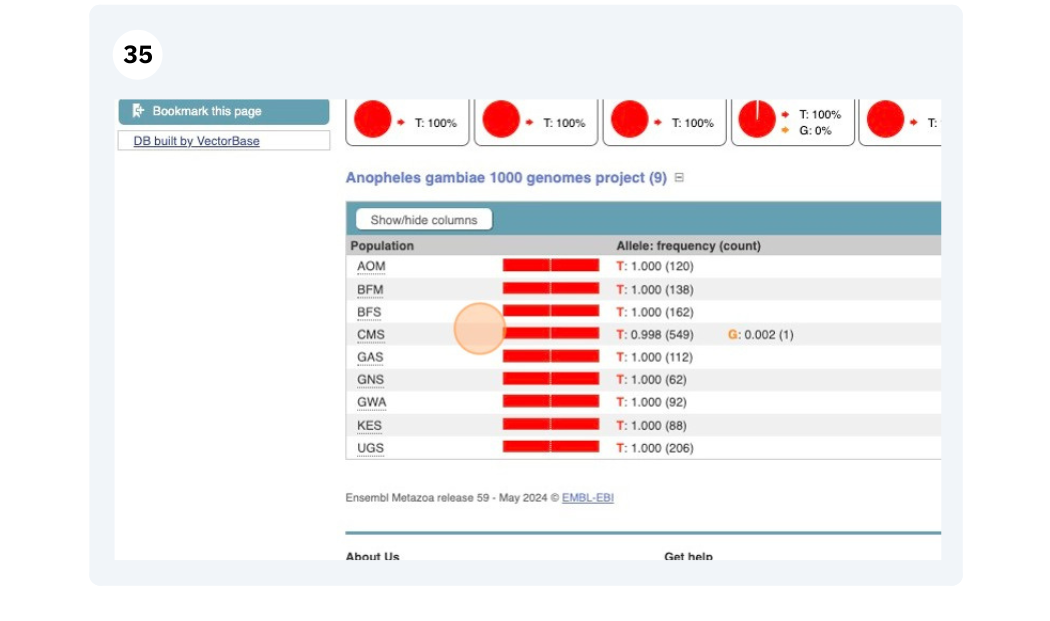
Exploring a SNP in Aedes aegypti
The Aedes aegypti AAEL004743 gene is part of ABC transporter superfamily associated with mosquito development and arboviral infections. Go to Ensembl Metazoa and answer the following questions:
-
How many variants have been identified in the gene that can cause a change in the protein sequence (i.e. missense variant)?
-
What is the ID of the variant that changes the amino acid residue 45 from Leucine to histidine (hint: refer to an amino acid codon table)? What is the location of this SNP in the Aedes aegypti (Yellow fever mosquito, LVP_AGWG) genome? What are its possible alleles?
-
Download the flanking sequence of this SNP in RTF (Rich Text Format). Can you change how much flanking sequence is displayed on the browser?
-
Does this SNP cause a change at the amino acid level for other genes or transcripts?
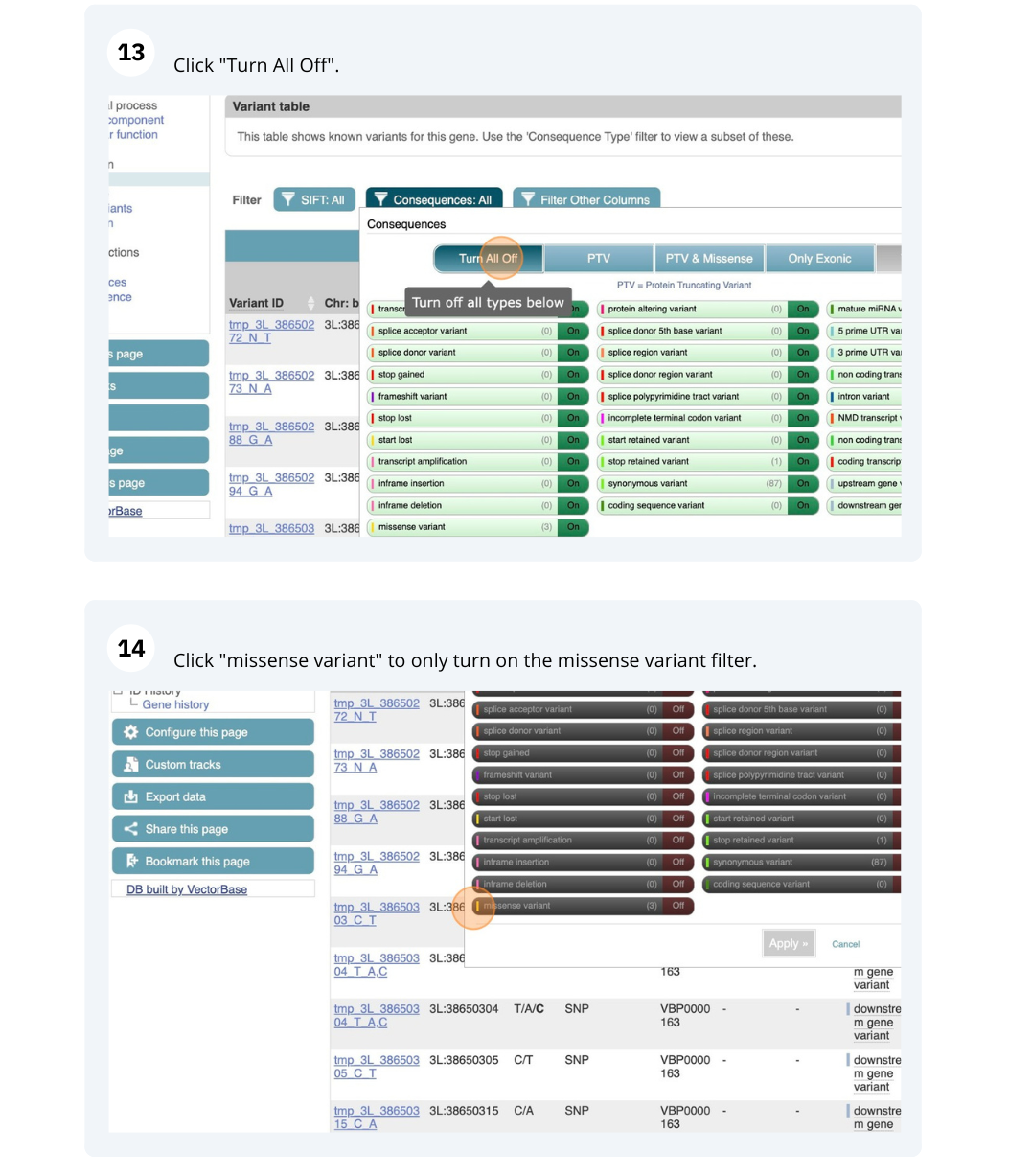
- Click on Aedes aegypti (Yellow fever mosquito, LVP_AGWG) on the Ensembl Metazoa homepage. Search for AAEL004743 on the species page. In the left-hand side menu of the Gene tab, click on Variant table. Click on Consequences: All then, Turn All off and select only missense variant.
The missense variant button indicates that there are 167 of these. Alternatively, you can count the number of variants in your filtered list.
- An amino acid codon table can be found on Wikipedia. Sort the AA coord column by clicking on the header and scroll down to find a variant at residue 45. The ID of this variant is supercont1.129:999463.
The variant is located at position 2:289218453. The two possible alleles at this locus are T (reference) and A (alternative).
- Click on the link supercont1.129:999463, then click on Flanking sequence in the left-hand side menu. Now click on Download sequence and select File format > Rich Text Format (RTF).
If you want to change how much flanking sequence is displayed on the browser, go back to the Flanking sequence page, click on the Configuration button and change the length of the sequence. The default settings is 400 bp.
- Click on Genes and regulation in the left-hand side menu.
This SNP is an upstream gene variant in AAEL004763 (A sequence variant located 5’ of a gene).
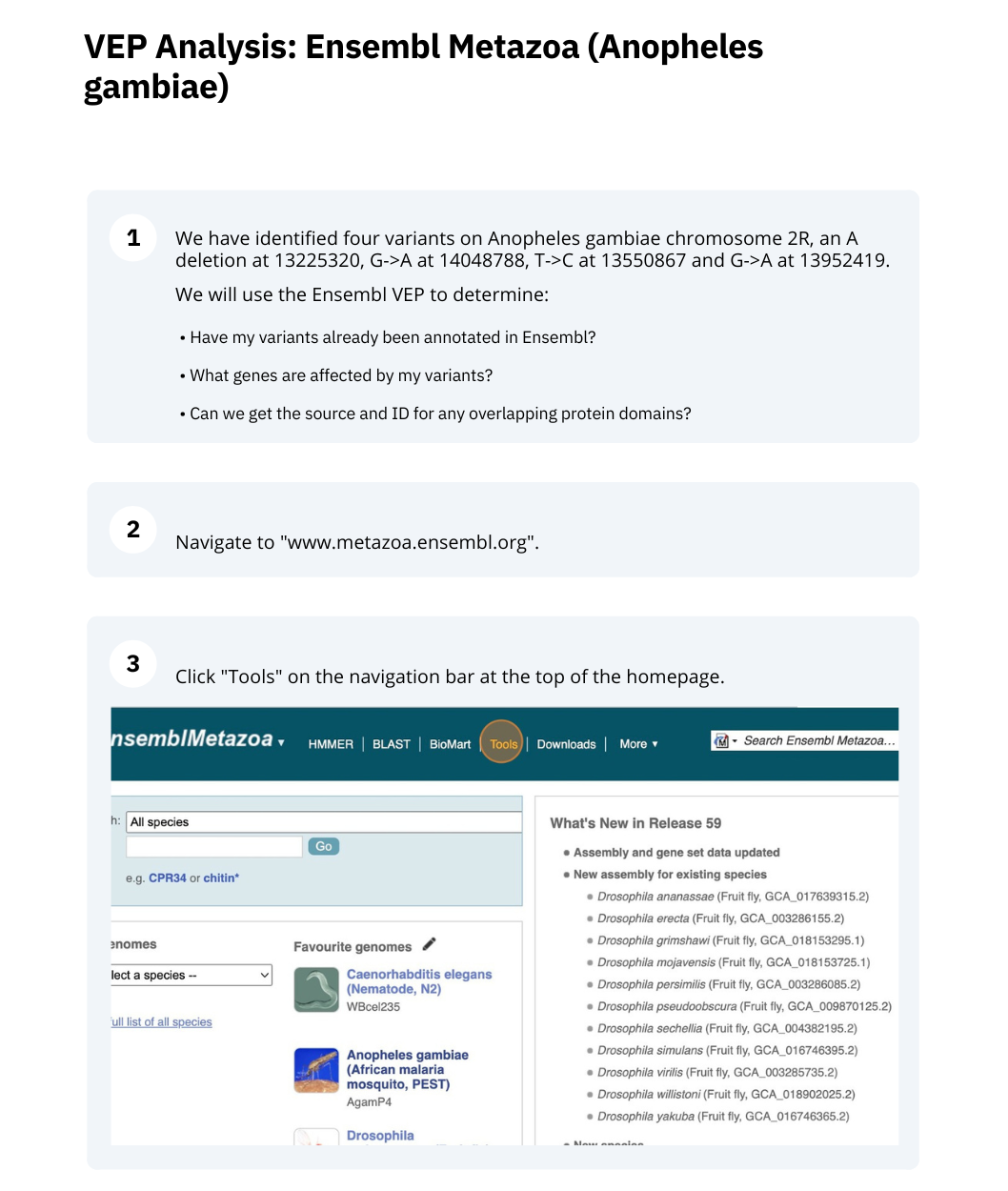
VEP
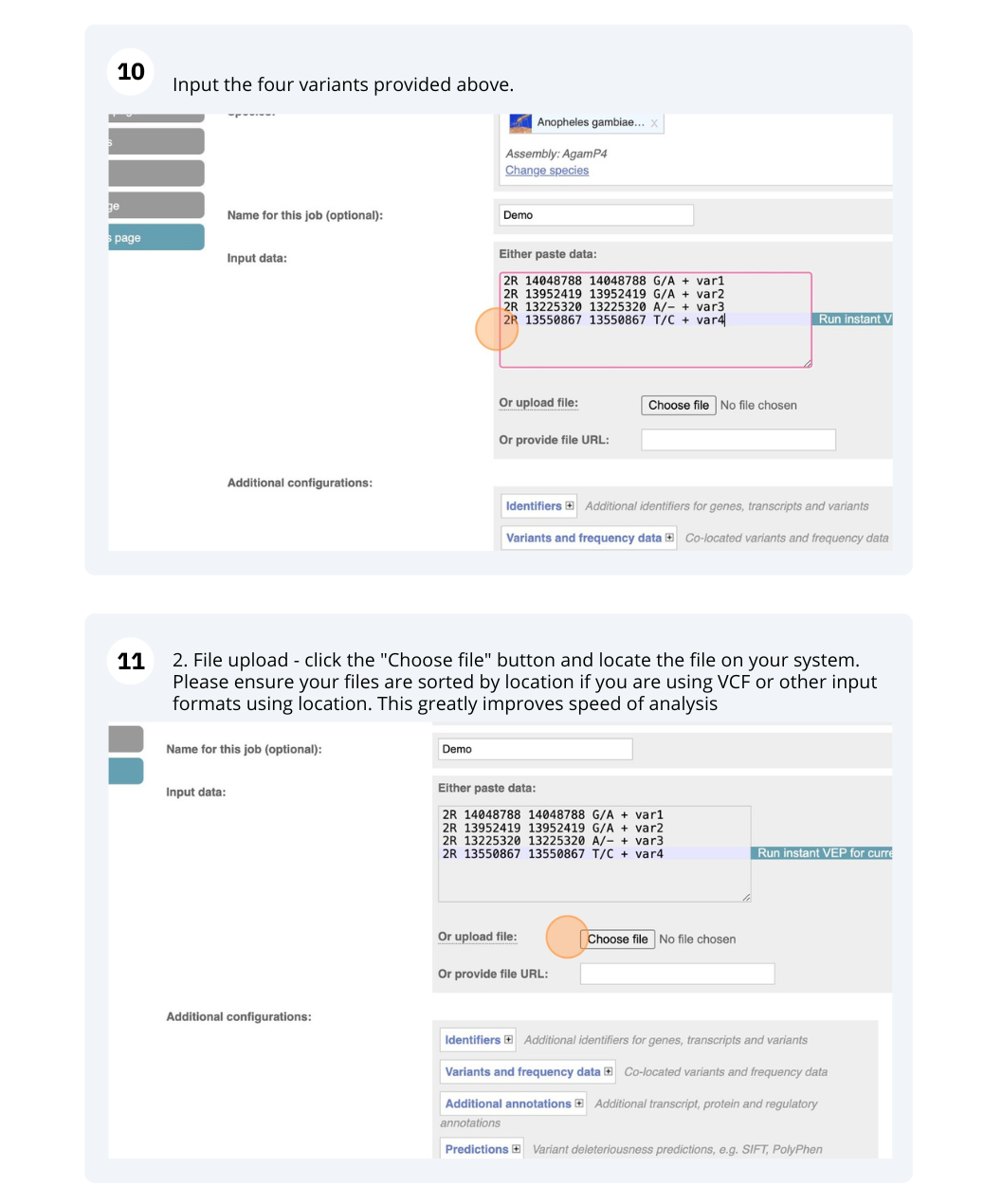
The data is in the format:
Chromosome Start End alleles (reference/alternative) strand name
Put the following into the Paste data box:
2R 14048788 14048788 G/A + var1
2R 13952419 13952419 G/A + var2
2R 13225320 13225320 A/- + var3
2R 13550867 13550867 T/C + var4
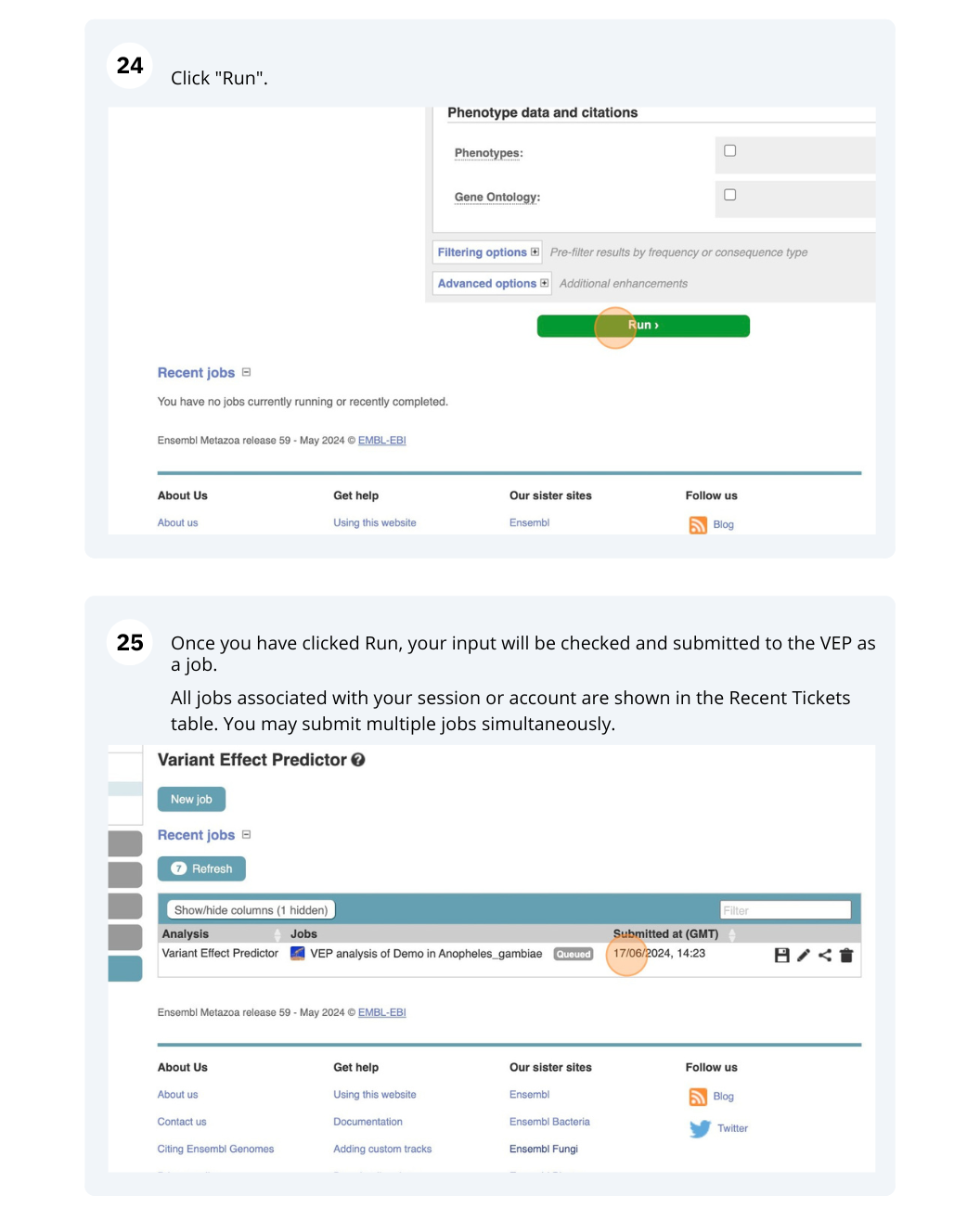
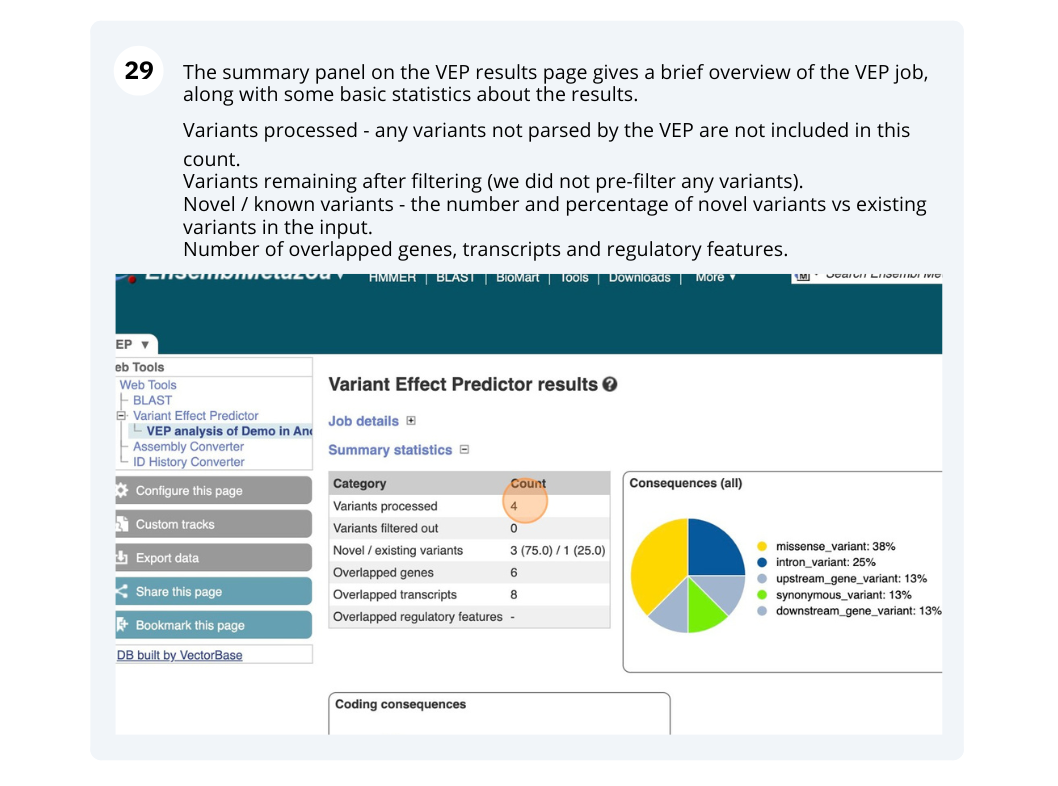
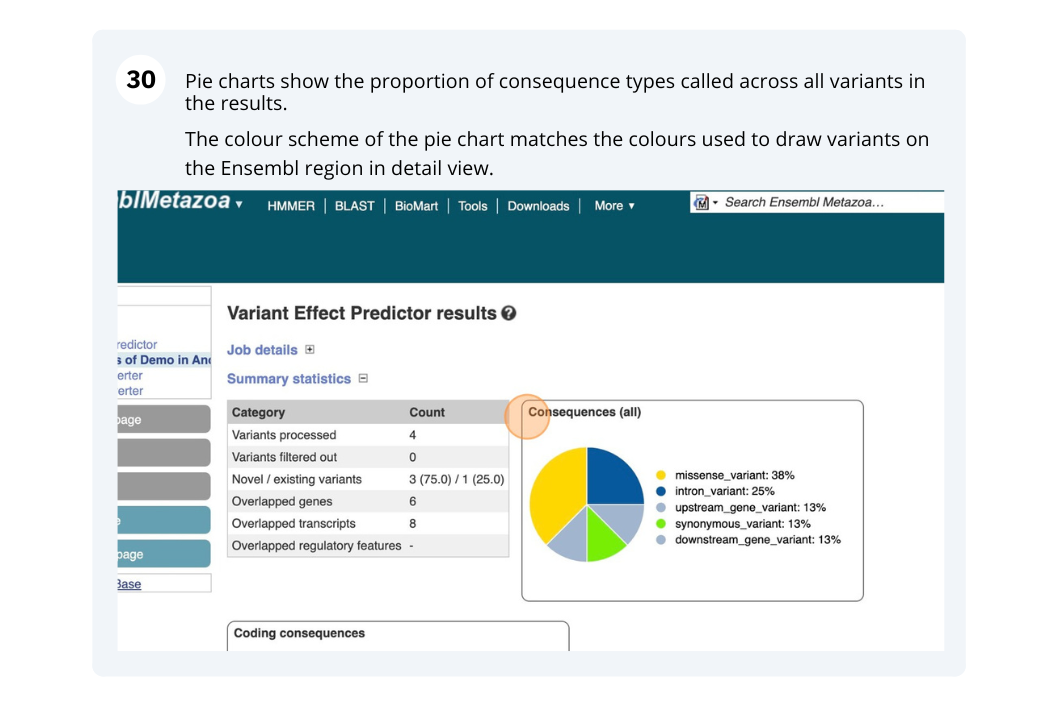
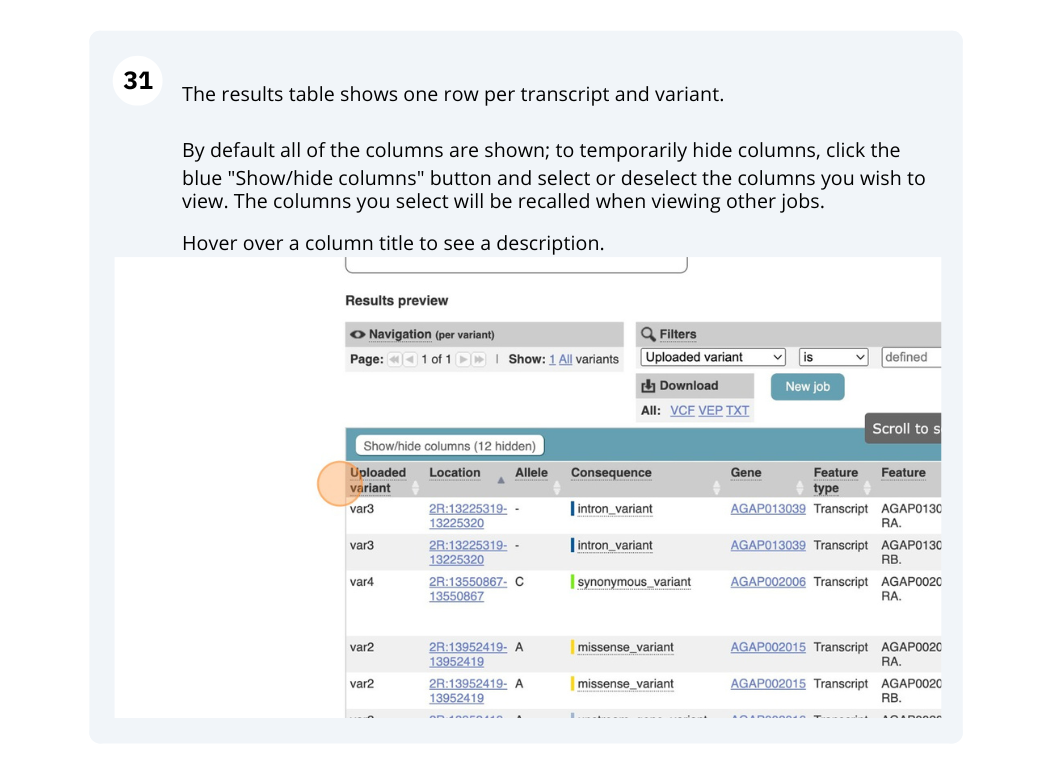
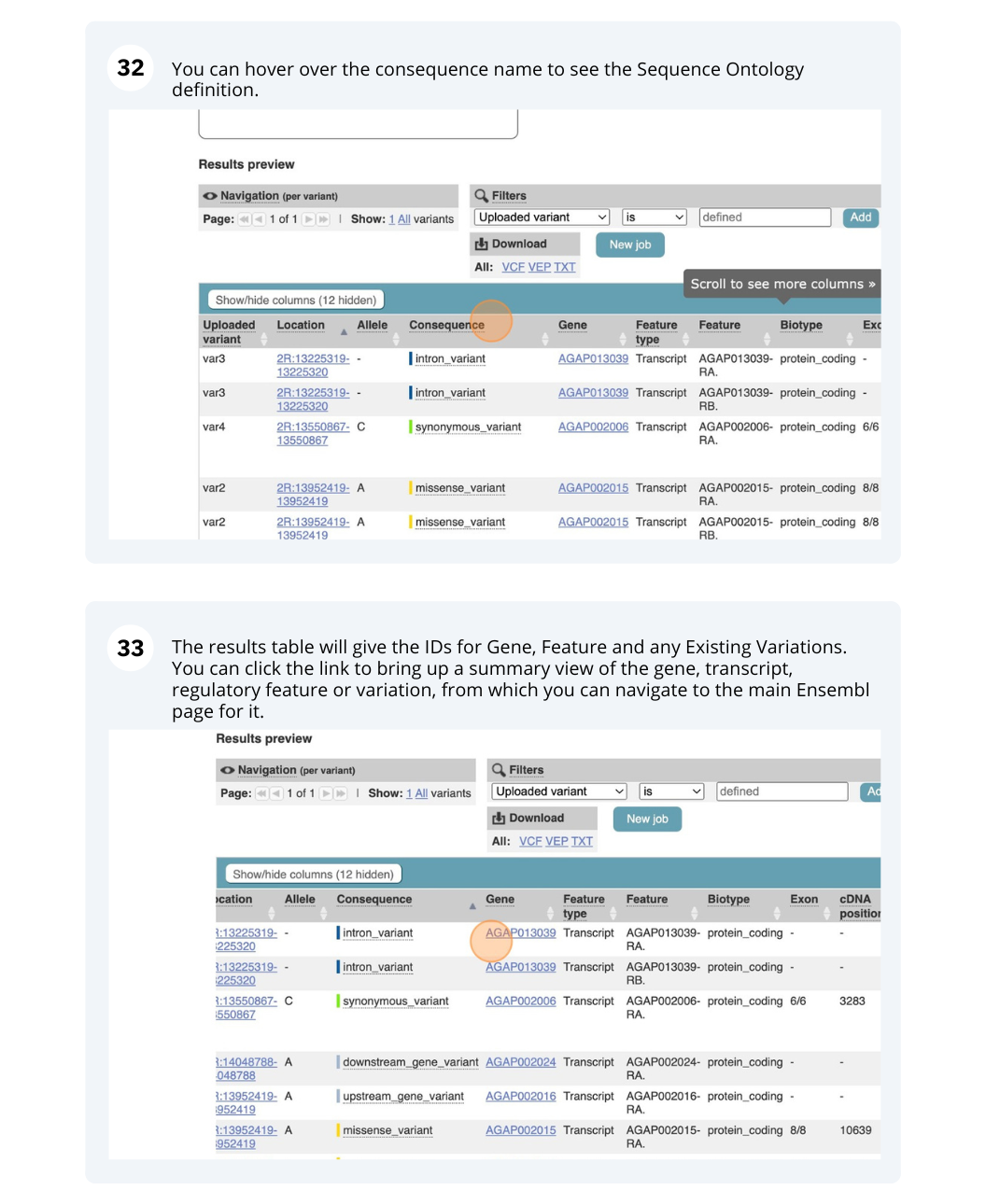
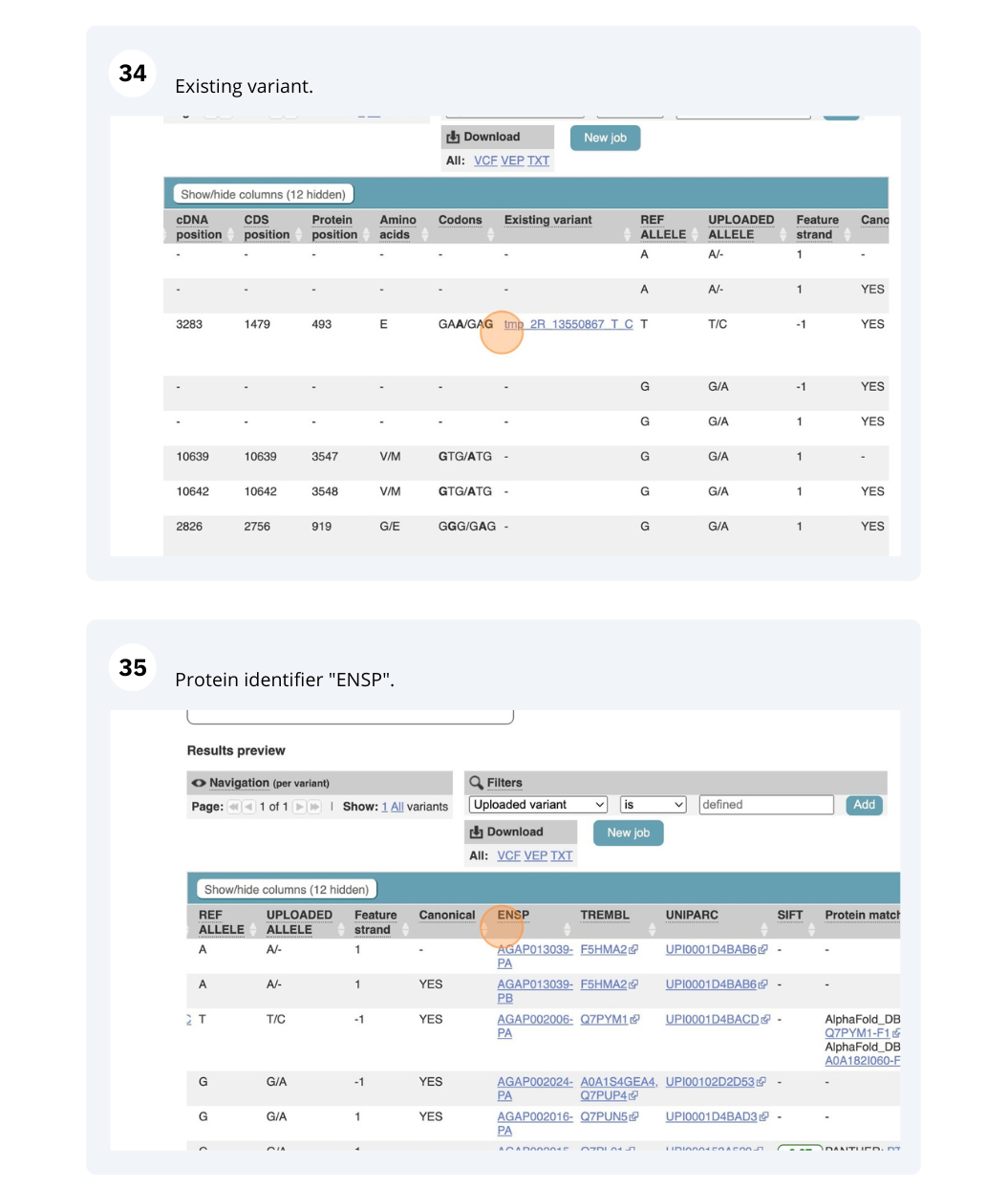
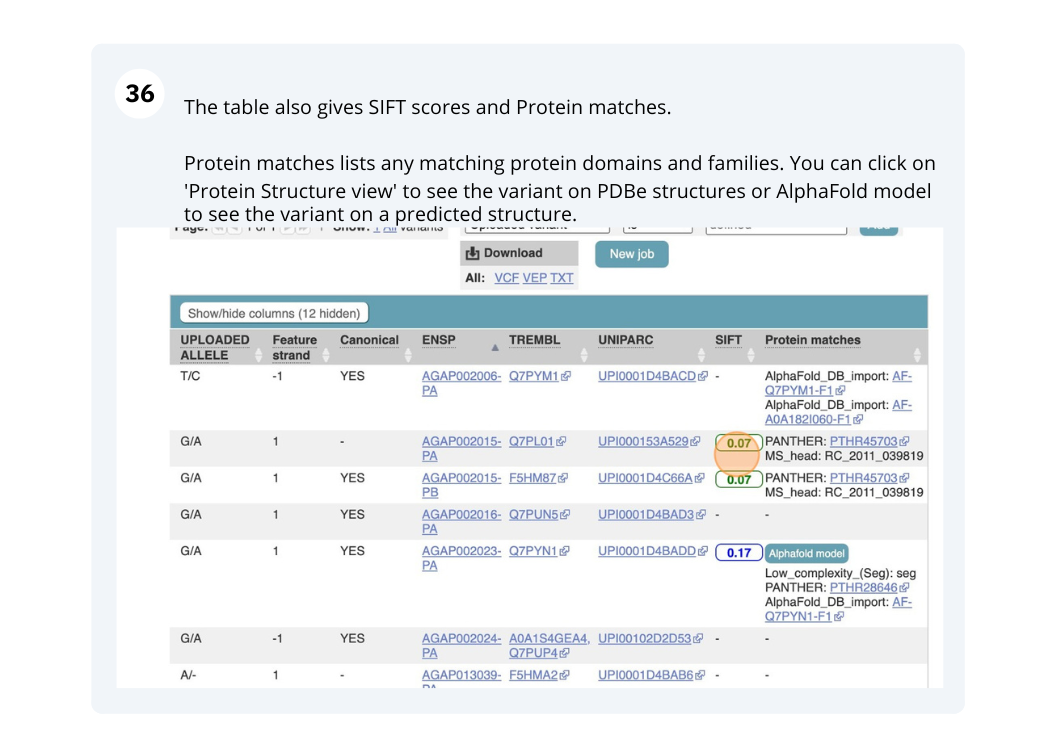
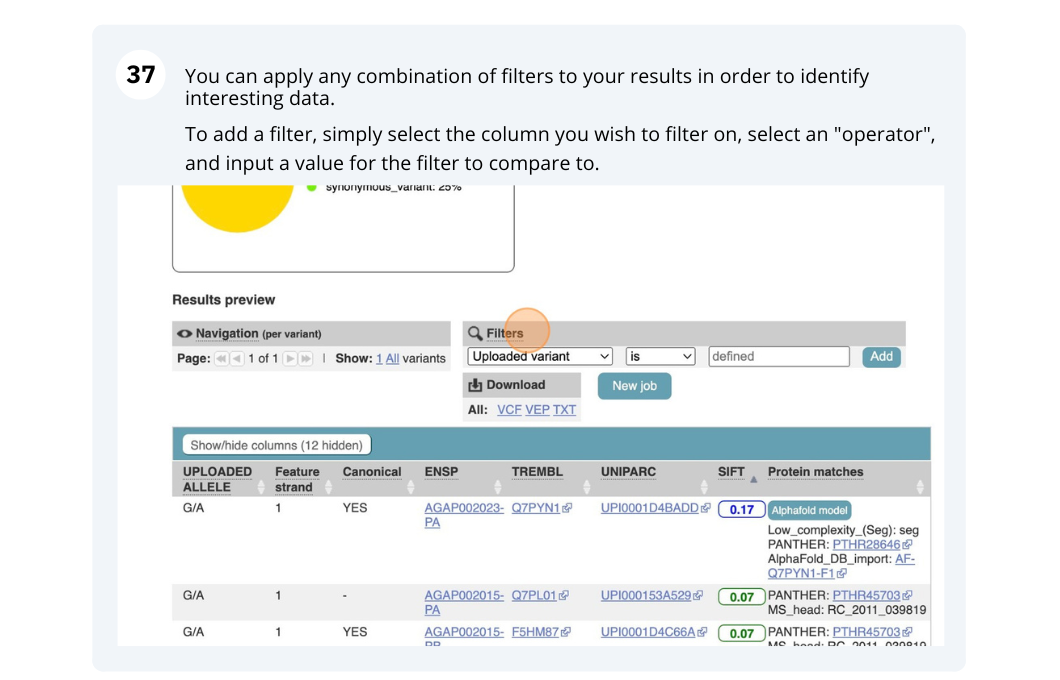
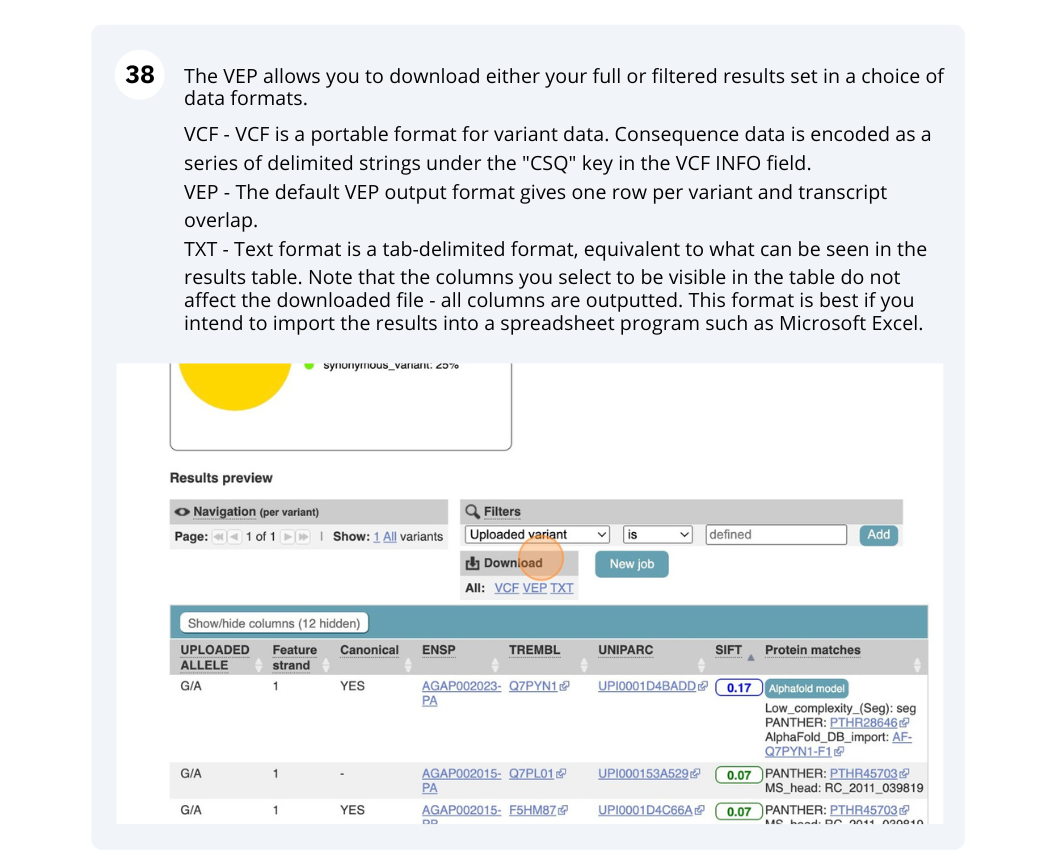
Running Anopheles gambiae variants through VEP
We have identified seven variants in Anopheles gambiae (African malaria mosquito, PEST).
tmp_X_2058015_T_C
tmp_X_2058017_G_C
tmp_3R_6014033_G_A
tmp_3R_6014046_A_T
tmp_3L_12676608_G_C
tmp_3L_12676741_A_G
tmp_2R_39524997_G_A
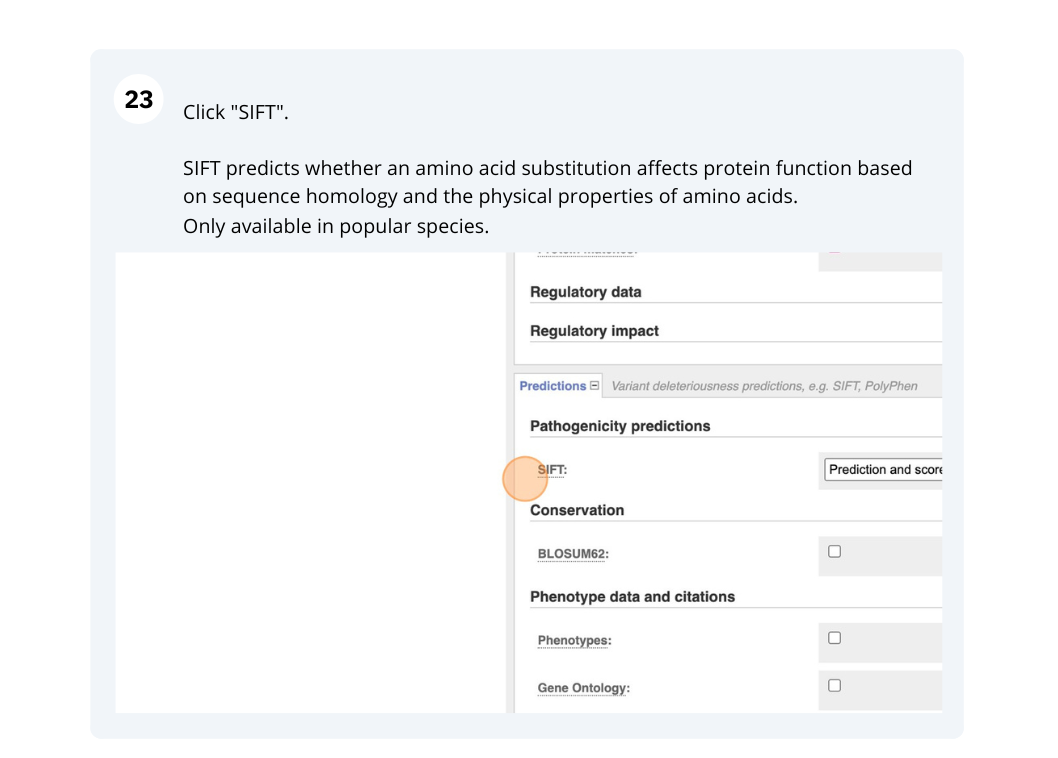
Use the VEP tool in Ensembl and choose the options to see SIFT predictions. Do these variants result in a change in the proteins encoded by any of the genes? Which gene? Have the variants already been found?
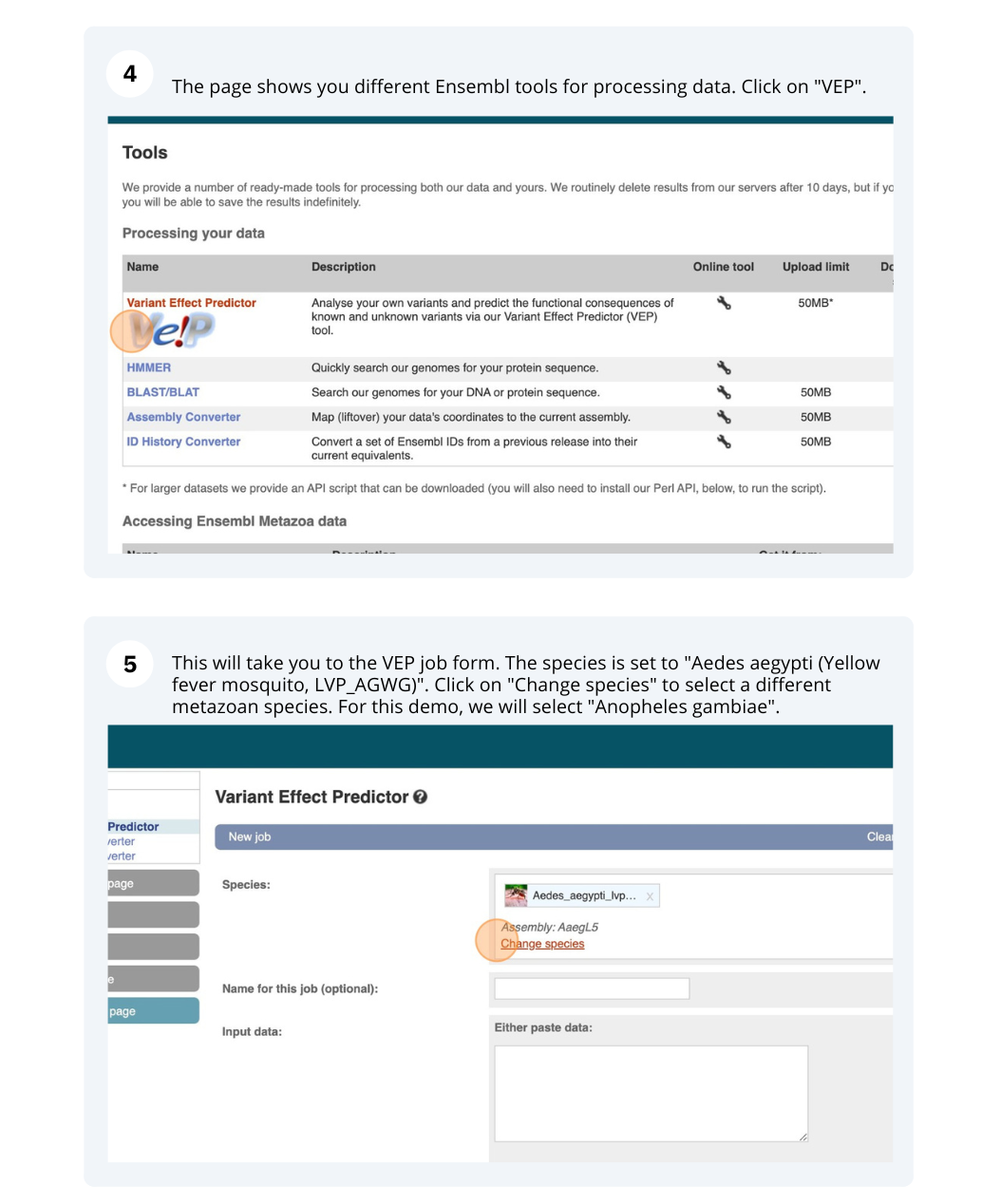
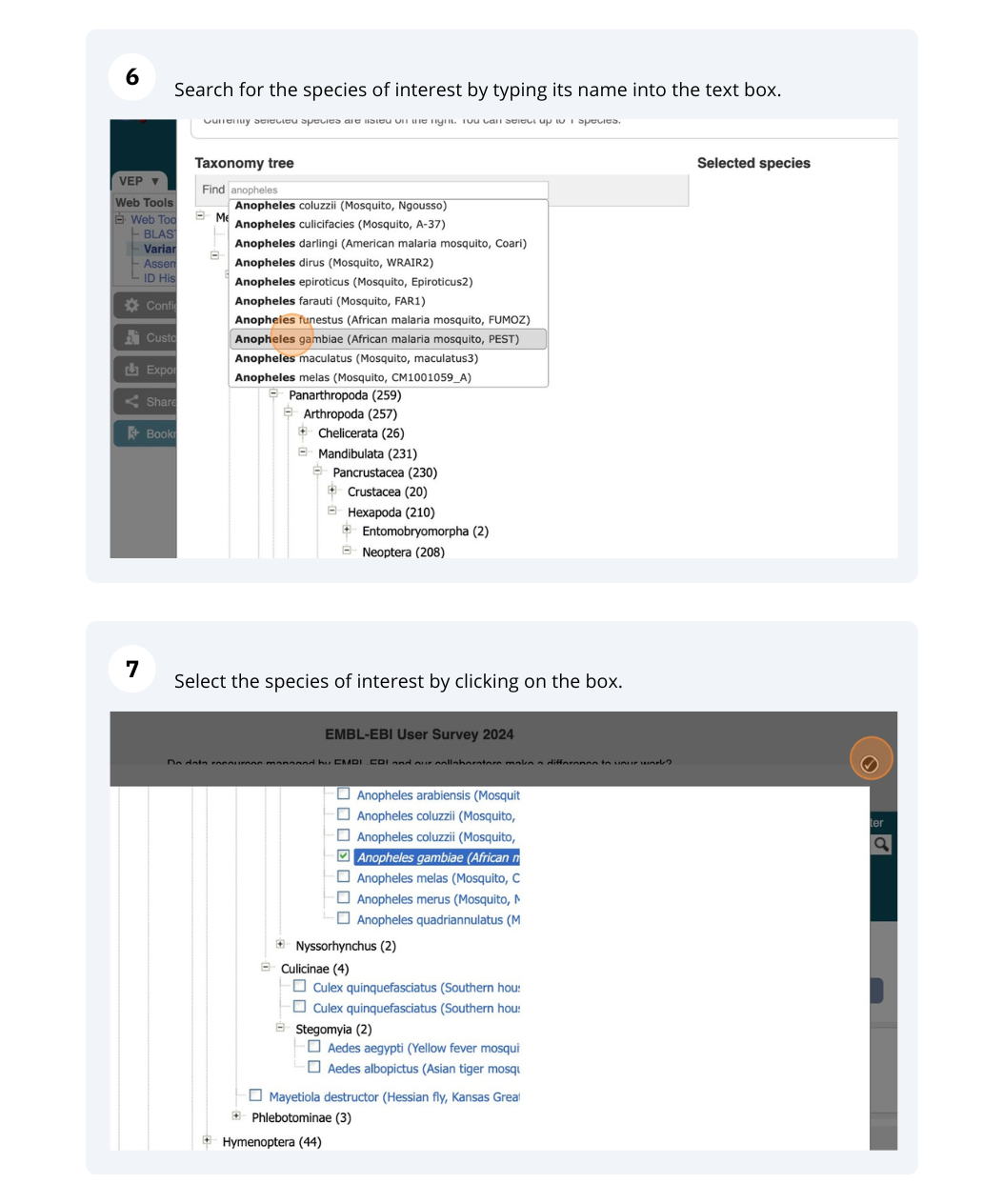
Go to the Ensembl Metazoa homepage and click on the link Tools at the top of the page. Click on Variant Effect Predictor and enter the variants in the text box:
Note: Variation data input can be done in a variety of formats. See more details about the different data formats and their structure in this VEP documentation page. Click Run. When your job is listed as Done, click View Results.
You will get a table with the consequence terms from the Sequence Ontology project (http://www.sequenceontology.org/) (i.e. synonymous, missense, downstream, intronic, 5’ UTR, 3’ UTR, etc) provided by VEP for the listed SNPs. You can also upload the VEP results as a track and view them on Location pages in Ensembl. SIFT is available for missense SNPs only.







