Filter Events by Year
Ensembl Browser Workshop: Plant & Animal Genome Conference (PAG) 30
Course Details
- Lead Trainer
- Louisse Paola Mirabueno
- Event Date
- 2023-01-15
- Location
- Town and Country D, Town & Country Resort and Conference Center, San Diego, CA 92108
- Description
- Work with the Ensembl Outreach team to get to grips with the Ensembl browser, accessing gene, variation, comparative genomics and regulation data, and mine these data with BioMart.
Demos and exercises
Species and genome assemblies
Demo: Introduction to Ensembl
Ensembl
Homepage
The front page of Ensembl is found at ensembl.org. It contains lots of information and links to help you navigate Ensembl:
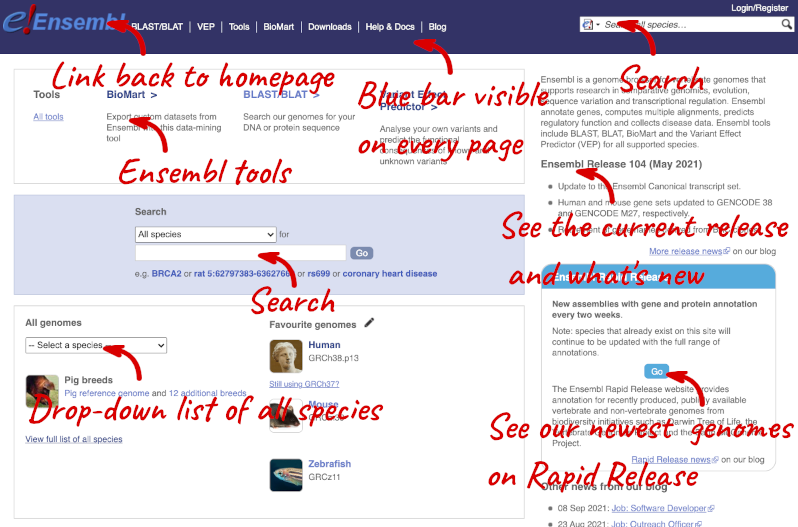
On the right-hand panel you can see the current release number and what has come out in this release. To access old releases, scroll to the bottom of the page and click on View in archive site in the right-hand corner.
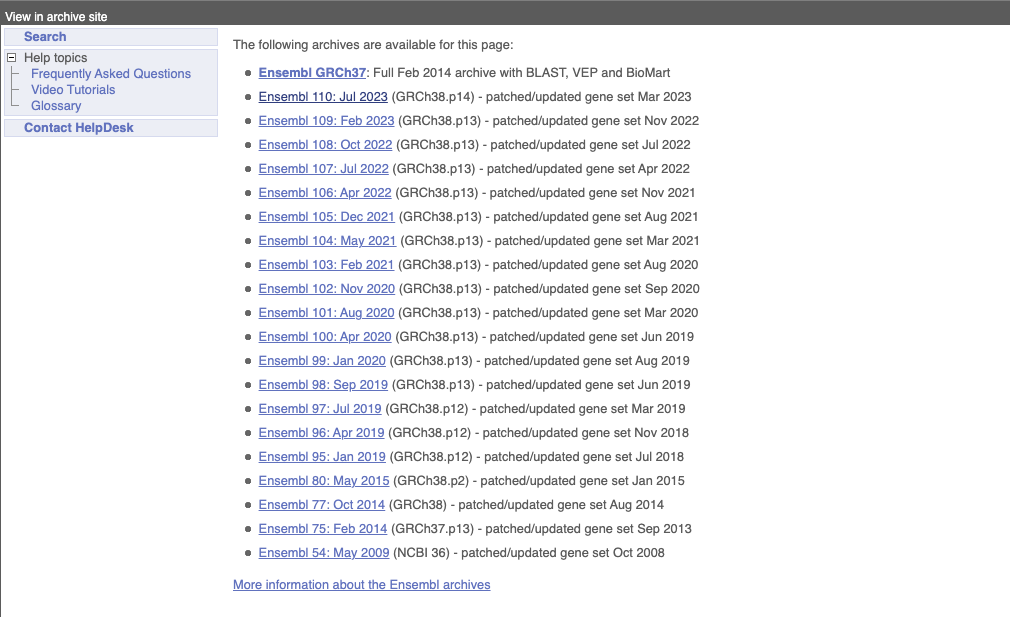
Click on the links to go to the archives. Alternatively, you can jump quickly to the correct release by adding e plus the release number in the URL. For example e98.ensembl.org jumps to Ensembl release 98.
Available species
Scroll back up to the top of the homepage. You can view all available species by clicking the View full list of all species link underneath the coloured search block.
You can search for your species of interest (either the common or scientific name) using the search bar at the top right-hand corner of the table. Click on the common name of your species of interest to go to the species information page. We’ll click on Human.
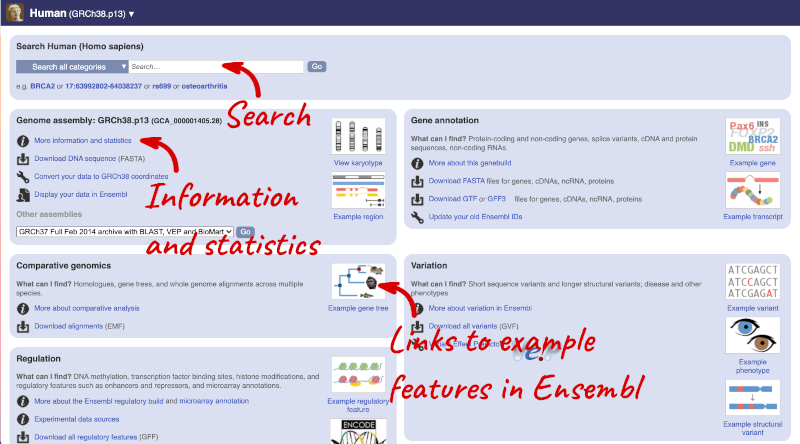
Species information
Here you can see links to example features and to download flatfiles. To find out more about the genome assembly and genebuild, click on More information and statistics under the Genome assembly section.
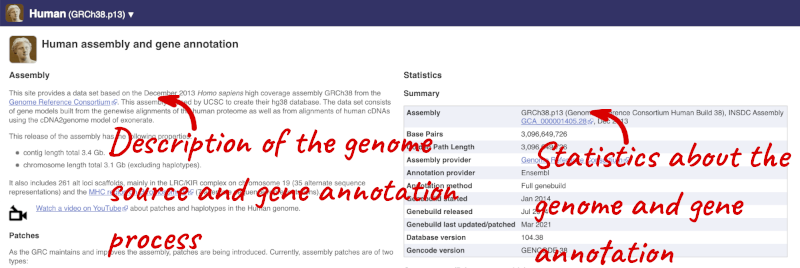
Here you’ll find a detailed description of how to the genome was produced and links to the original source. You will also see details of how the genes were annotated.
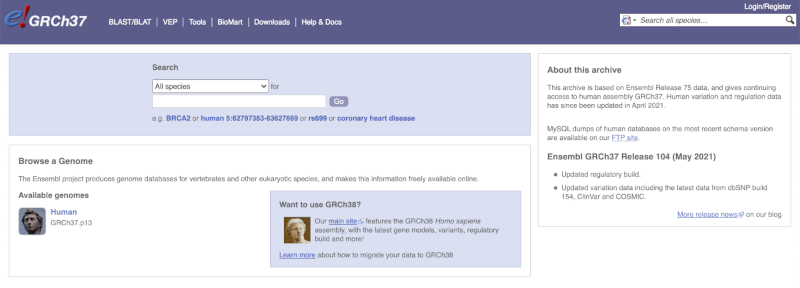
The current genome assembly for human is GRCh38. If you want to see the previous assembly, GRCh37, visit our dedicated site grch37.ensembl.org.
Ensembl Genomes
Homepage
Let’s take a look at the Ensembl Genomes homepage at ensemblgenomes.org.
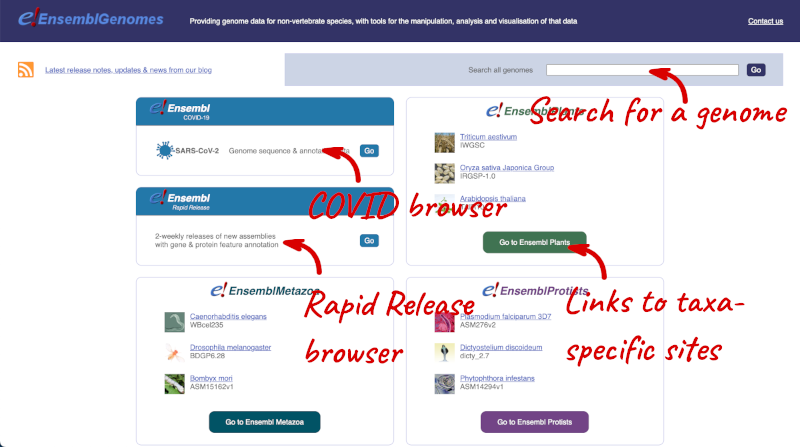
Click on the different taxa to see their homepages. Each one has a different colour-coding, but they are all structured in a similar format to the Ensembl main site.
You can navigate most of the taxa in the same way as you would with Ensembl.
Ensembl Bacteria
Ensembl Bacteria has a large number of genomes and has a slightly different method to the other Ensembl sites. Let’s look at it in more detail.
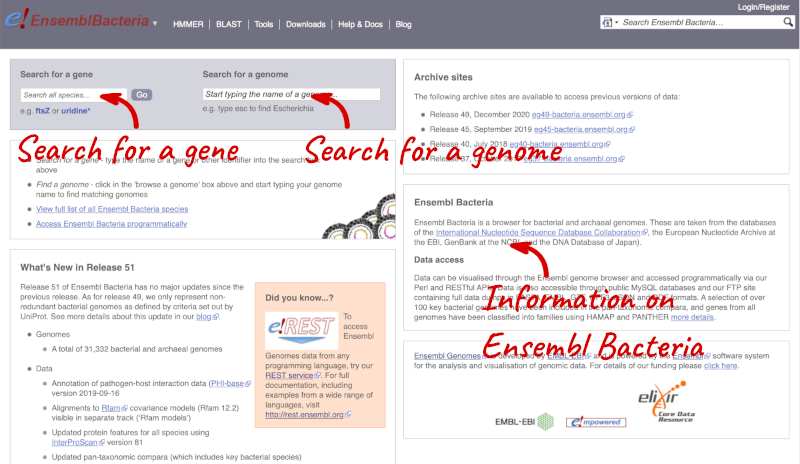
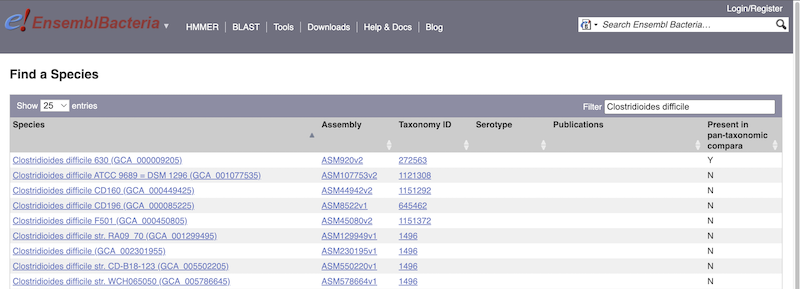
There’s no drop-down species list for bacteria as it would be hard to navigate with the number of species. You can click the View full list of all Ensembl Bacteria species link underneath the coloured search block. Search for your species of interest using the filter in the top right-hand corner of the table.
Alternatively, you can find a species by typing the species name into the Search for a genome search box at the top of the page. A drop-down list will appear with any species matching the name you entered.
For example, to find a sub-strain of Clostridioides difficile start typing in the species name. Due to the auto-complete, you’ll see useful results as soon as you get to Clostridio.
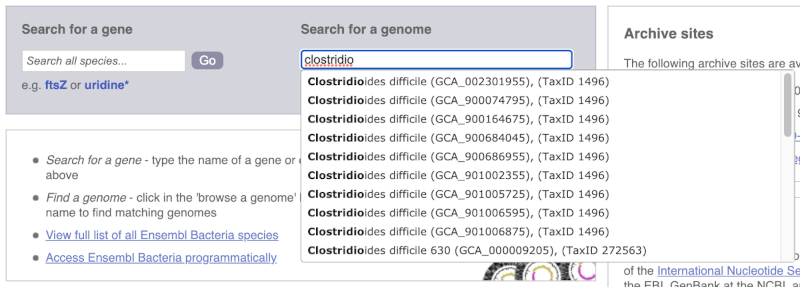
The drop down contains various strains of C. difficile. Let’s choose C. difficile 630. This will take us to another species information page, where we can explore various features.
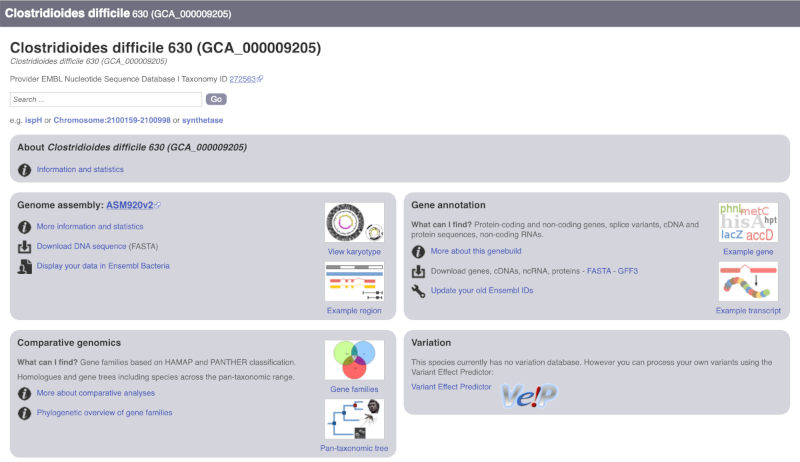
Unlike the Homo sapiens species information page, there is no prose description of the genome or gene annotation, as these pages were generated automatically.
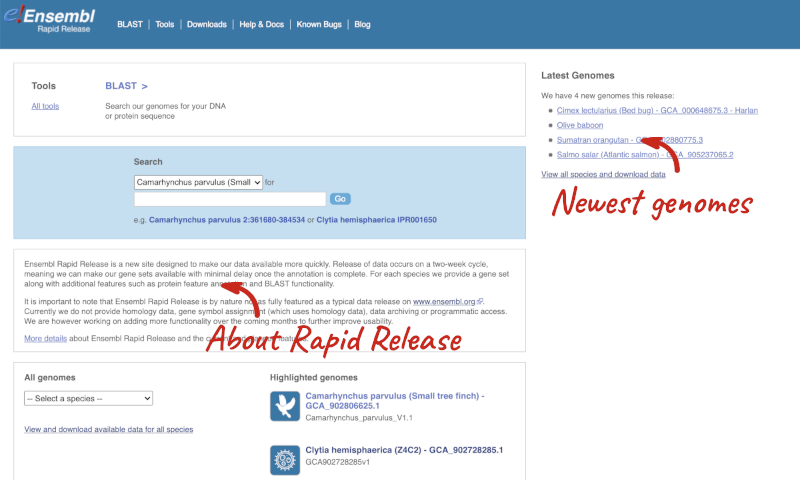
Ensembl Rapid Release
Our newest genomes, such as those coming from the Darwin Tree of Life, are available rapid.ensembl.org with limited annotation.
Genes and Transcripts
VEP
Demonstration of the VEP web interface
Input
We have identified three variants on wheat chromosome 4B:
C -> T at 240206468
C -> G at 240199078
C -> T at 240212229
We will use the Ensembl VEP to determine:
- Have my variants already been annotated in Ensembl?
- What genes are affected by my variants?
- Do any of my variants affect gene regulation?
Click on Tools in the top green bar from any Ensembl Plants page, then Variant Effect Predictor to open the input form:
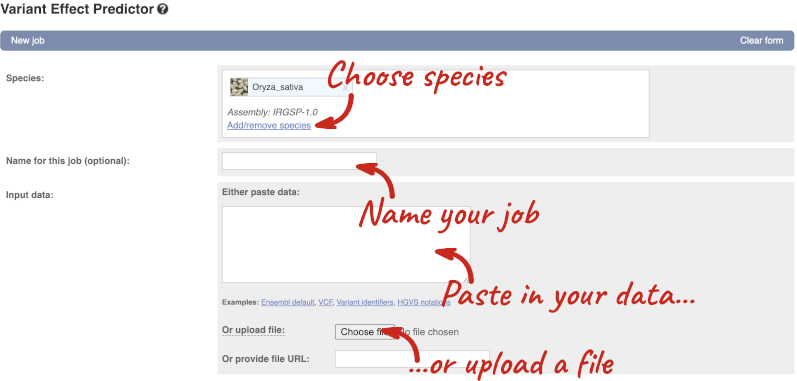
Click on Add/remove species and search for Triticum aestivum to choose it.
The data is in VCF:
chromosome coordinate id reference alternative
Put the following into the Paste data box:
4B 240206468 var1 C T
4B 240199078 var2 C G
4B 240212229 var3 C T
The VEP will automatically detect that the data is in VCF.
Additional configurations
There are further options that you can choose for your output. These are categorised as Identifiers, Variants and frequency data, Additional annotations, Predictions, Filtering options and Advanced options. Let’s open all the menus and take a look.
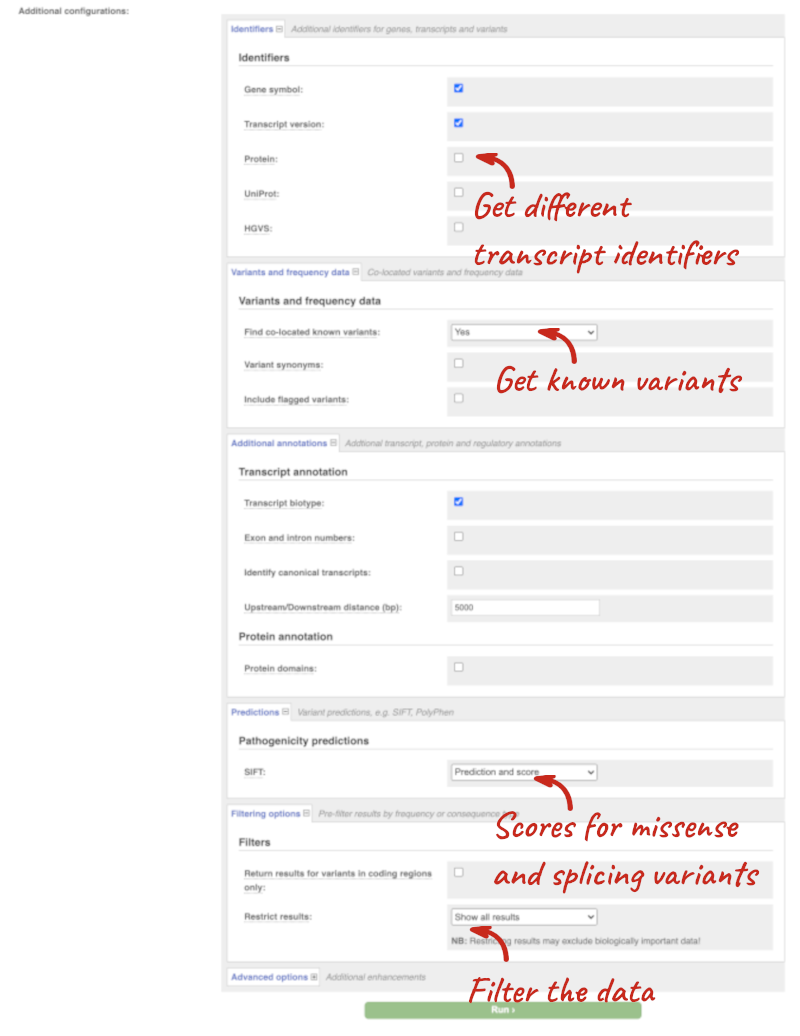
Hover over the options to see definitions.
When you’ve selected everything you need, scroll right to the bottom and click Run.

The display will show you the status of your job. It will say Queued, then automatically switch to Done when the job is done, you do not need to refresh the page. You can edit or discard your job at this time. If you have submitted multiple jobs, they will all appear here.
Click View results once your job is done.
Results
In your results you will see a graphical summary of your data, as well as a table of your results.
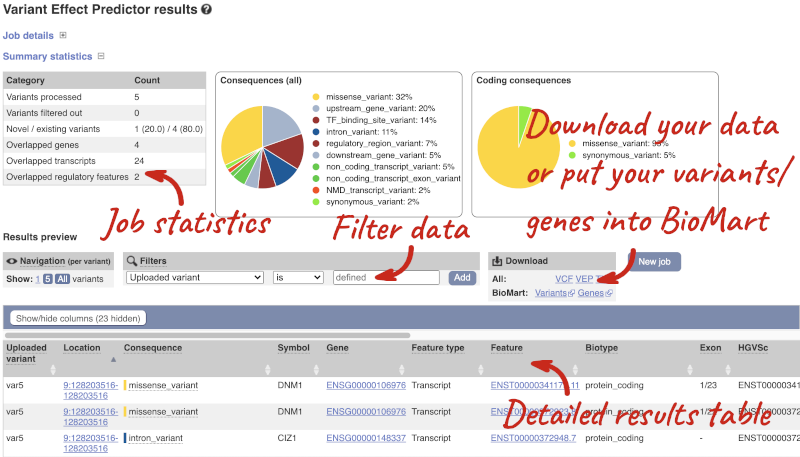
The results table is enormous and detailed, so we’re going to go through the it by section. The first column is Uploaded variant. If your input data contains IDs, like ours does, the ID is listed here. If your input data is only loci, this column will contain the locus and alleles of the variant. You’ll notice that the variants are not neccessarily in the order they were in in your input. You’ll also see that there are multiple lines in the table for each variant, with each line representing one transcript or other feature the variant affects.
You can mouse over any column name to get a definition of what is shown.
The next few columns give the information about the feature the variant affects, including the consequence. Where the feature is a transcript, you will see the gene symbol and stable ID and the transcript stable ID and biotype. The IDs are links to take you to the gene or transcript homepage.
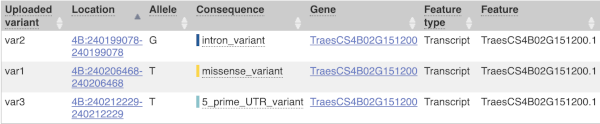
This is followed by details on the effects on transcripts, including the position of the variant in terms of the exon number, cDNA, CDS and protein, the amino acid and codon change and pathogenicity scores. Where the variant is known, the ID of the existing variant is listed, with a link out to the variant homepage. The pathogenicity scores are shown as numbers with coloured highlights to indicate the prediction, and you can mouse-over the scores to get the prediction in words.
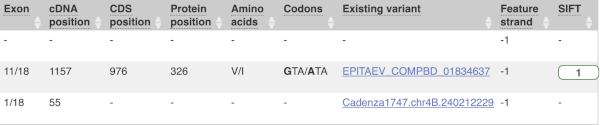
Above the table is the Filter option, which allows you to filter by any column in the table. You can select a column from the drop-down, then a logic option from the next drop-down, then type in your filter to the following box. We’ll try a filter of Consequence, followed by is then missense_variant, which will give us only variants that change the amino acid sequence of the protein. You’ll notice that as you type missense_variant, the VEP will make suggestions for an autocomplete.
You can export your VEP results in various formats, including VCF. When you export as VCF, you’ll get all the VEP annotation listed under CSQ in the INFO column. After filtering your data, you’ll see that you have the option to export only the filtered data. You can also drop all the genes you’ve found into the Gene BioMart, or all the known variants into the Variation BioMart to export further information about them.
BioMart
Follow these instructions to guide you through BioMart to answer the following query:
You have three questions about a set of sheep genes:
ESPN, MYH9, USH1C, CISD2, THRB
(these are VGNC gene symbols. More details on the Vertebrate Gene Nomenclature Committee can be found on https://vertebrate.genenames.org/)
- What are the NCBI Gene IDs for these genes?
- Are there associated functions from the GO (gene ontology) project that might help describe their function?
- What are their cDNA sequences?
Click on BioMart in the top header of a www.ensembl.org page to go to: www.ensembl.org/biomart/martview
You cannot choose any filters or attributes until you’ve chosen your dataset. Your dataset is the data type you’re working with. In this case we’re going to choose sheep genes, so pick Ensembl Genes then Sheep genes from the drop-downs.
Now that you’ve chosen your dataset, the filters and attributes will appear in the column on the left. You can pick these in any order and the options you pick will appear.
Click on Filters on the left to see the available filters appear on the main page. You’ll see that there are loads of categories of Filters to choose from. You can expand these by clicking on them. For our query, we’re going to expand GENE.
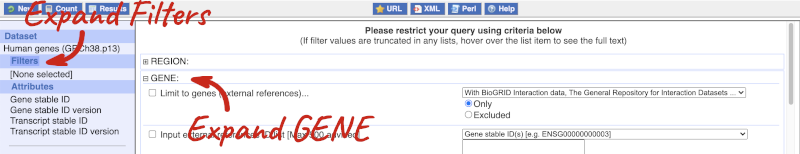
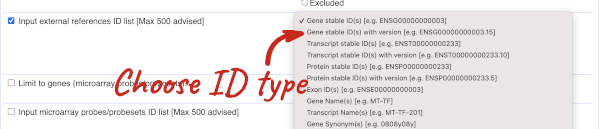
Our input data is a list of identifiers, so we’re going to use the Input external references ID list filter. This allows us to input a list of identifiers from different databases. We need to choose what kind of identifier we’re using, so that BioMart can look up the right column in a data table. You can pick these from a drop-down list, which lists the type of identifier with an example of how it looks. For our query, we have a list of gene names, so we need to pick Gene Name(s).
To check if the filters have worked, you can use the Count button at the top left, which will show you how many genes have passed the filter. If you get 0 or another number you don’t expect, this can help you to see if your query was effective.
To choose the attributes, expand this in the menu. There are five categories for sheep gene attributes. These categories are mutually exclusive, you cannot pick attributes from multiple categories. This means that we need to do two separate queries to get our GO terms and NCBI IDs, and to get our cDNA sequences.

The Ensembl gene and transcript IDs, with and without version numbers are selected by default. The selected attributes are also listed on the left.

We can choose the attributes we want by clicking on them. For our query, we’re going to select:
- GENE
- Gene Name
- EXTERNAL
- NCBI gene ID
- GO term accession
- GO term name
- GO term definition
We need to select the Gene Name in order to get back our original input, as this is not returned by default in BioMart. The order that you select the attributes in will define the order that the columns appear in in your output table.
You can get your results by clicking on Results at the top left.

The results table just gives you a preview of the first ten lines of your query. This allows the results to load quickly, so that if you need to make any changes to your query, you don’t waste any time. To see the full table you can click on View ## rows. You can also export the data to an xls, tsv, csv or html file. For large queries, it is recommended that you export your data as Compressed web file (notify by email), to ensure your download is not disrupted by connection issues.
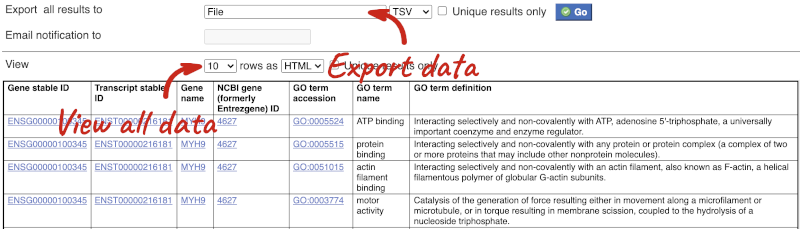
You can see multiple rows per gene in your input list, because there are multiple transcripts per gene and multiple GO terms per transcript.
To get the cDNA sequences, go back to the Attributes then select the category Sequences and expand SEQUENCES.
When you select the sequence type, the part of the transcript model you’ve chosen will be highlighted in the grpahic.
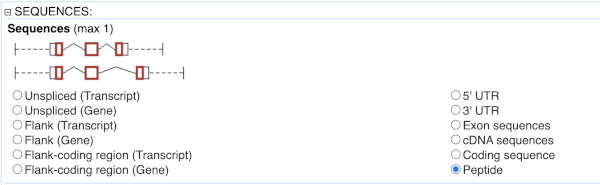
Choose cDNA sequences, then expand HEADER INFORMATION to add Gene Name to the header. Then hit Results again.

For more details on BioMart, have a look at this publication:
Kinsella, R.J. et al
Ensembl BioMarts: a hub for data retrieval across taxonomic space.
http://europepmc.org/articles/PMC3170168







