Filter Events by Year
Ensembl Plants Browser Workshop: Designing Future Wheat (DFW)
Course Details
- Lead Trainer
- Louisse Paola Mirabueno
- Event Dates
- 2023-12-14 until 2023-12-15
- Location
- John Innes Centre, Norwich, UK
- Description
- Work with the Ensembl Outreach team to get to grips with the Ensembl Plants browser.
Demos and exercises
Species and genome assemblies
Demo: Exploring species and genome assemblies in Ensembl Plants
Homepage
The front page of Ensembl Plants is found at plants.ensembl.org. It contains lots of information and links to help you navigate Ensembl Plants:
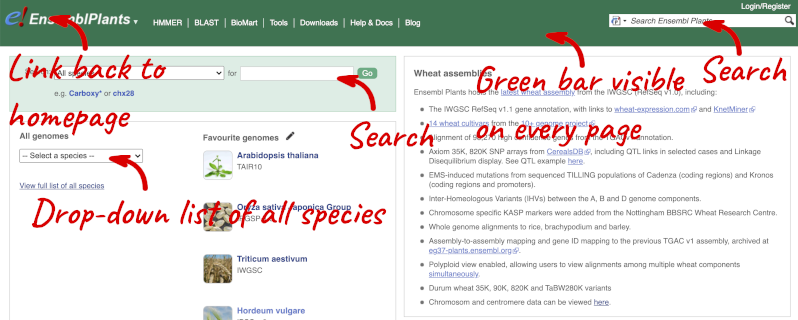
At the top left you can see the current release number and what has come out in this release.
Available species
Click on View full list of all species.
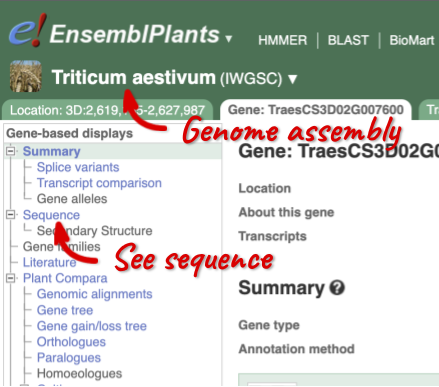
Click on the scientific name of your species of interest to go to the species homepage. We’ll click on Triticum aestivum.
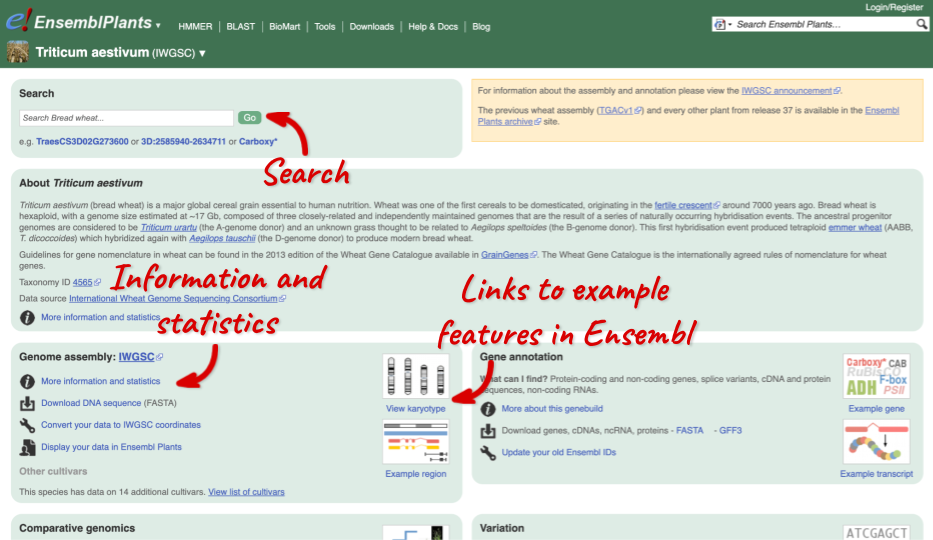
Species information
Here you can see links to example pages and to download flatfiles. To find out more about the genome assembly and genebuild, click on More information and statistics.
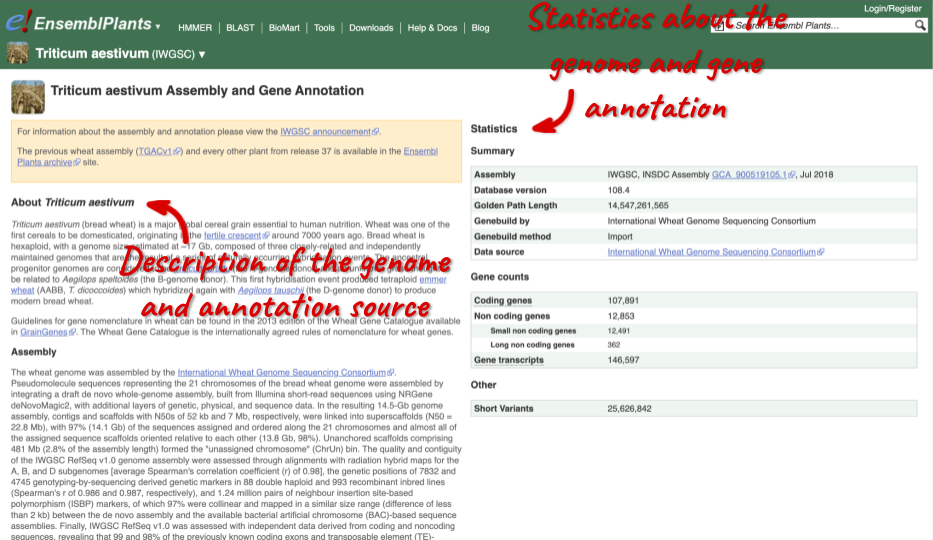
Here you’ll find a detailed description of how to the genome was produced and links to the original source. You will also see details of how the genes were annotated.
Triticum aestivum (wheat) cultivars
-
Are there any additional cultivars available alongside the Triticum aestivum (IWGSC) reference genome?
-
Find the description of the wheat assembly. Which institute provided the assembly and annotations?
-
How many coding and non-coding genes does the IWGSC assembly have?
-
Are there any other species of the genus Triticum available in Ensembl? If so, which species are they?
- Go to Ensembl Plants and click on Triticum aestivum on the front page of Ensembl Plants to go to the species information page. Under the Genome assembly section of the species page, you will find the number of cultivars in wheat.
There are 14 cultivars.
- Click on More information and statistics in the Genome assembly section and scroll down to the paragraph on Assembly.
The assembly and annotations were generated by the International Wheat Genome Sequencing Consortium (IWGSC).
- Stay on the More information and statistics page. You can find some summary statistics on the right-hand side.
The T. aestivum (IWGSC) assembly has 107,891 coding and 12,853 non-coding genes.
- Go to the Ensembl Plants homepage. Click on View full list of all species in the All genomes panel. Filter the table by entering
Triticumin the text box on the top right-hand corner of the table.Besides T. aestivum are 4 other Triticum species available in Ensembl: Triticum dicoccoides (wild emmer wheat), Triticum spelta (spelt), Triticum turgidum (domesticated emmer wheat) and Triticum urartu (red wild einkorn wheat).
Exploring genomic regions
Demo: Exploring genomic regions in Ensembl Plants
Start at the Ensembl Plants front page. You can search for a region by typing it into a search box, but you have to specify the species.
To bypass the text search, you need to input your region coordinates in the correct format, which is chromosome, colon, start coordinate, dash, end coordinate, with no spaces for example: 1D:41289600-41345600. Choose Triticum aestivum from the species drop-down, then type (or copy and paste) these coordinates into the search box.
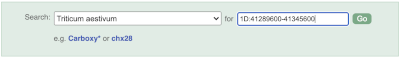
Press Enter or click Go to jump directly to the Region in detail Page.
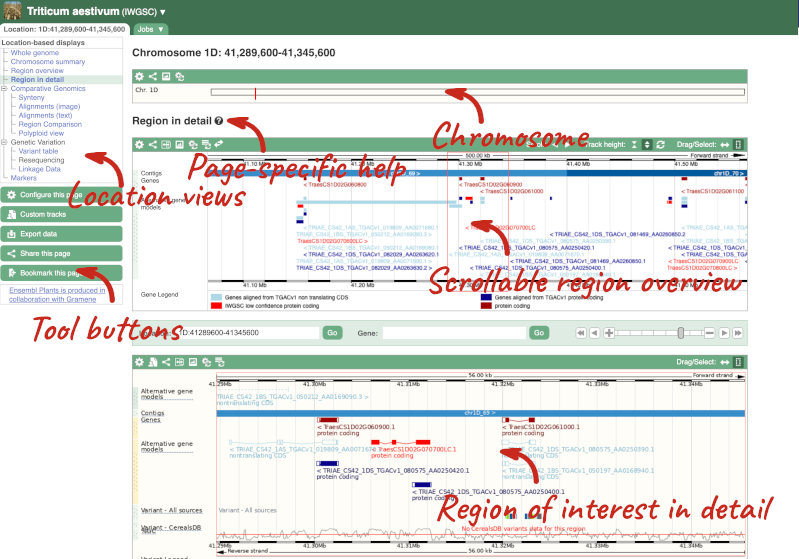
Click on the  button to view page-specific help. The help pages provide text, labelled images and, in some cases, help videos to describe what you can see on the page and how to interact with it.
button to view page-specific help. The help pages provide text, labelled images and, in some cases, help videos to describe what you can see on the page and how to interact with it.
The Region in detail page is made up of three images, let’s look at each one in detail.
- The first image shows the chromosome:

The region we’re looking at is highlighted on the chromosome. You can jump to a different region by dragging out a box in this image. Drag out a box on the chromosome, a pop-up menu will appear.
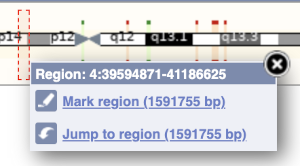
If you wanted to move to the region, you could click on Jump to region (### bp). If you wanted to highlight it, click on Mark region (###bp). For now, we’ll close the pop-up by clicking on the X in the corner.
- The second image shows a 1Mb region around our selected region. This is always 1Mb in human, but the fixed size of this view varies between species. This view allows you to scroll back and forth along the chromosome.
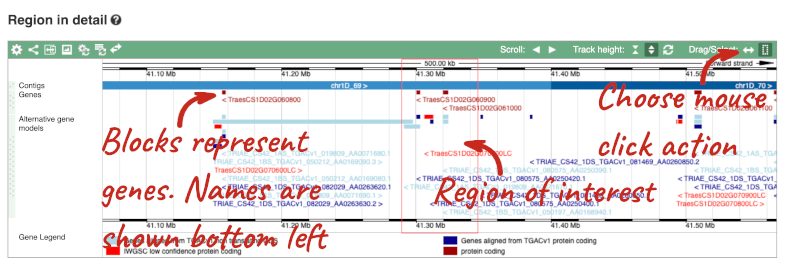
You can also drag out and jump to or mark a region.
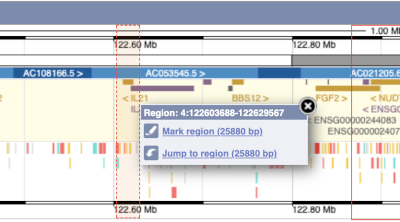
Click on the X to close the pop-up menu.
Click on the Drag/Select button  to change the action of your mouse click. Now you can scroll along the chromosome by clicking and dragging within the image. As you do this you’ll see the image below grey out and two blue buttons appear. Clicking on Update this image would jump the lower image to the region central to the scrollable image. We want to go back to where we started, so we’ll click on Reset scrollable image.
to change the action of your mouse click. Now you can scroll along the chromosome by clicking and dragging within the image. As you do this you’ll see the image below grey out and two blue buttons appear. Clicking on Update this image would jump the lower image to the region central to the scrollable image. We want to go back to where we started, so we’ll click on Reset scrollable image.

- The third image is a detailed, configurable view of the region.

Here you can see various tracks, which is what we call a data type that you can plot against the genome. Some tracks, such as the transcripts, can be on the forward or reverse strand. Forward stranded features are shown above the blue contig track that runs across the middle of the image, with reverse stranded features below the contig. Other tracks, such as variants, regulatory features or conserved regions, refer to both strands of the genome, and these are shown by default at the very top or very bottom of the view.
You can use click and drag to either navigate around the region or highlight regions of interest, Click on the Drag/Select option at the top or bottom right to switch mouse action. On Drag, you can click and drag left or right to move along the genome, the page will reload when you drop the mouse button. On Select you can drag out a box to highlight or zoom in on a region of interest.
With the tool set to Select, drag out a box around an exon and choose Mark region.
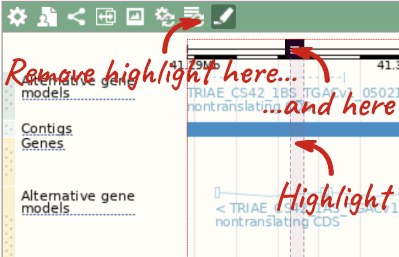
The highlight will remain in place if you zoom in and out or move around the region. This allows you to keep track of regions or features of interest.
We can edit what we see on this page by clicking on the blue Configure this page menu at the left.

This will open a menu that allows you to change the image.
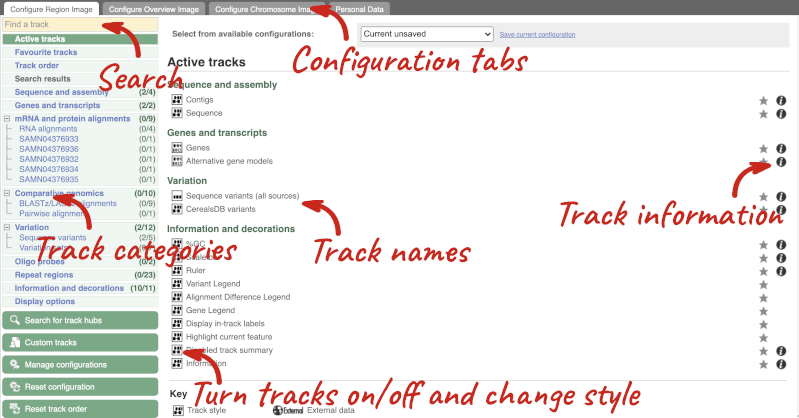
There are thousands of possible tracks that you can add. When you launch the view, you will see all the tracks that are currently turned on with their names on the left and an info icon on the right, which you can click on to expand the description of the track. Turn them on or off, or change the track style by clicking on the box next to the name. More details about the different track styles are in this FAQ.
You can find more tracks to add by either exploring the categories on the left, or using the Find a track option at the top left. Type in a word or phrase to find tracks with it in the track name or description.
Let’s add some tracks to this image. Add:
- EMS-induced mutation variants
- Type I Transposons/LINE (Repeats: Repbase)
Now click on the tick in the top left hand to save and close the menu. Alternatively, click anywhere outside of the menu. We can now see the tracks in the image.
If the track is not giving you can information you need, you can easily change the way the tracks appear by hovering over the track name then the cog wheel to open a menu. To make it easier to compare information between tracks, such as spotting overlaps, you can move tracks around by clicking and dragging on the bar to the left of the track name.

Now that you’ve got the view how you want it, you might like to show something you’ve found to a colleague or collaborator. Click on the Share this page button to generate a link. Email the link to someone else, so that they can see the same view as you, including all the tracks you’ve added. These links contain the Ensembl release number, so if a new release or even assembly comes out, your link will just take you to the archive site for the release it was made on.

To return this to the default view, go to Configure this page and select Reset configuration at the bottom of the menu.
Due to hybridisations in wheat’s evolutionary history, it has a hexaploid genome with related homoeologous regions. We can compare these with the Polyploid view. First, let’s zoom in on the gene TraesCS1D02G061000 by dragging out a box around it and clicking on Jump to region. Now click on the Polyploid view link in the left-hand menu.
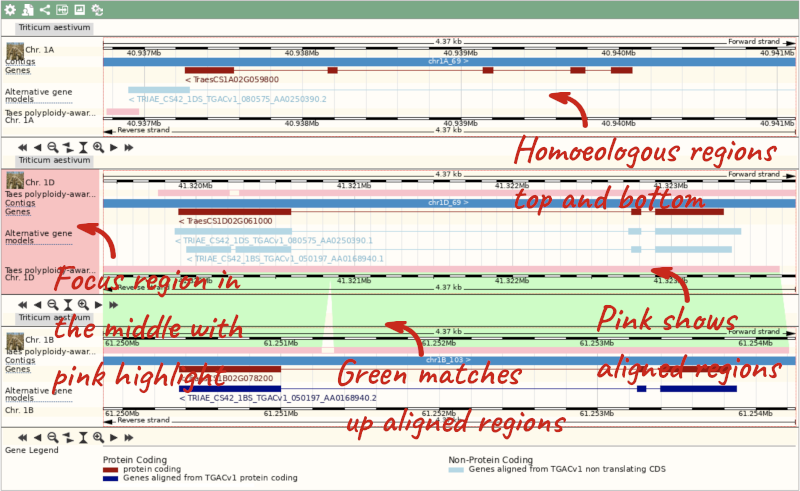
This view also allows us to configure the page, as we could with the main region view, so that we can compare other features between the homoeologous chromosomes.
Exploring a wheat region
-
Go to 2D:378720500-378780600 in Triticum aestivum (wheat).
-
How many genes are in this region? What strand are the genes on? What are the gene IDs for these genes?
-
What tracks can you see that show gene structure? Where did the different tracks come from?
-
Export the genomic sequence for this region.
-
Can you view the genomic alignments of the homoeologous regions? What are the different formats you can export the image as?
-
Go to the Ensembl Plants homepage. Select Search: Triticum aestivum and type
2D:378720500-378780600in the text box. Click Go. -
There are two genes displayed in the Genes track. They are both located on the reverse strand. The IDs are
-
There are two tracks which have mapping to this gene: Genes and Alternative gene models. Click the track names for more information on their source.
-
Click Export data in the left-hand menu. Leave the default parameters as they are. Click Next>. Click on Text. Note that the sequence has a header that provides information about the genome assembly, the chromosome, the start and end coordinates and the strand. For example:
>2D dna:chromosome chromosome:IWGSC:2D:378720500:378780600:1 -
Click on Polyploid view in the left hand menu to view the homoeologous regions. Click on Export image. This will open a pop-up menu of the different image formats you can export, which are PNG and PDF.
Genes and transcripts
Demo: Viewing genes and transcripts
You can find out lots of information about Ensembl genes and transcripts using the browser. If you’re already looking at a region view, you can click on any transcript and a pop-up menu will appear, allowing you to jump directly to that gene or transcript.

Alternatively, you can find a gene by searching for it. You can search for gene names or identifiers, and also phenotypes or functions that might be associated with the genes.
We’re going to look at the Triticum aestivum TraesCS3D02G007600 gene. From the Ensembl Plants homepage, type TraesCS3D02G007600 into the search bar and click the Go button.
The gene tab
Click on TraesCS3D02G007600 from the search hits. The Gene tab should open:
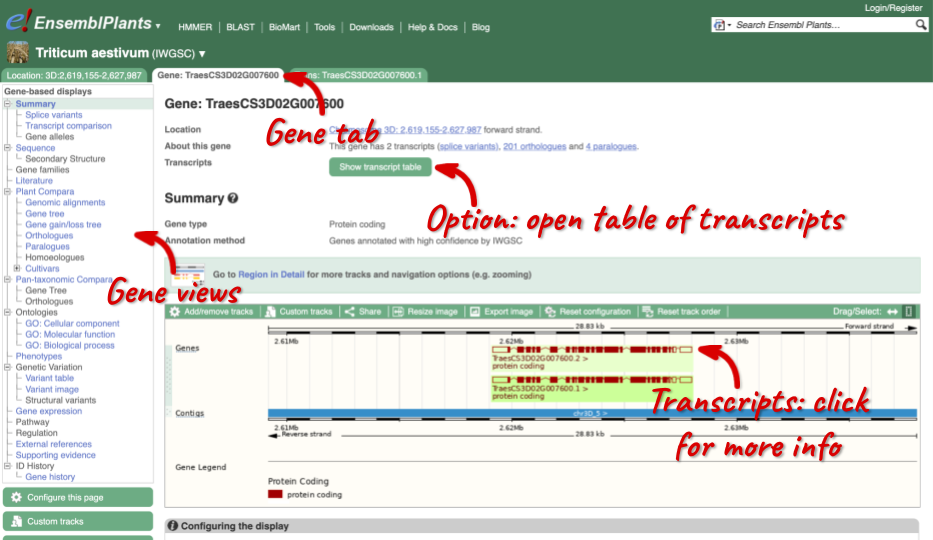
This page summarises the gene, including its location, name and equivalents in other databases. At the bottom of the page, a graphic shows a region view with the transcripts. We can see exons shown as blocks with introns as lines linking them together. Coding exons are filled, whereas non-coding exons are empty. We can also see the overlapping and neighbouring genes and other genomic features.
There are different tabs for different types of features, such as genes, transcripts or variants. These appear side-by-side across the blue bar, allowing you to jump back and forth between features of interest. Each tab has its own navigation column down the left hand side of the page, listing all the things you can see for this feature.
Gene sequence
Let’s walk through this menu for the gene tab. How can we view the genomic sequence? Click Sequence at the left of the page.
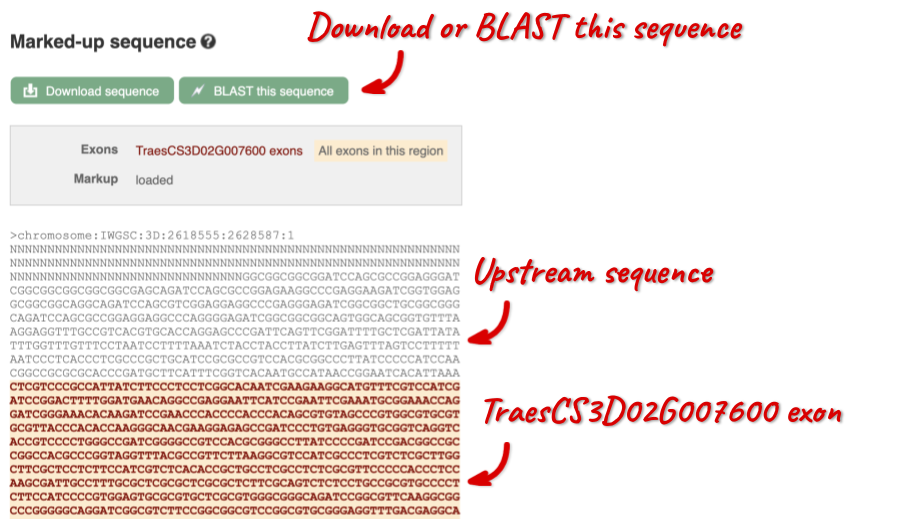
The sequence is shown in FASTA format. The FASTA header contains the genome assembly, chromosome, coordinates and strand (1 or -1) – this gene is on the positive strand.
Exons are highlighted within the genomic sequence, both exons of our gene of interest and any neighbouring or overlapping gene. By default, 600 bases are shown up and downstream of the gene. We can make changes to how this sequence appears with the blue Configure this page button found at the left. This allows us to change the flanking regions, add variants, add line numbering and more. Click on it now.
Once you have selected changes (in this example, Show variants and Line numbering) click at the top right.

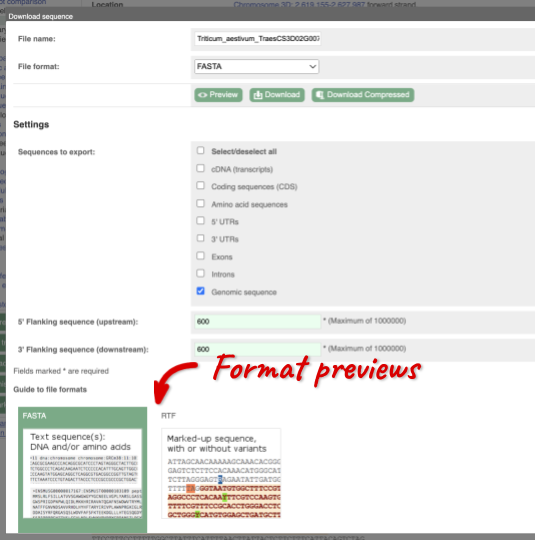
You can download this sequence by clicking in the Download sequence button above the sequence:
This will open a dialogue box that allows you to pick between plain FASTA sequence, or sequence in rich-text format (RTF), which includes all the coloured annotations and can be opened in a word processor. If you want run a sequence analysis tool, download as FASTA sequence, whereas if you want to analyse the sequence visually, RTF is best for this. This button is available for all sequence views.
Gene function
To find out what the protein does, have a look at GO terms from the Gene Ontology consortium. There are three pages of GO terms, representing the three divisions in GO: Biological process (what the protein does), Cellular component (where the protein is) and Molecular function (how it does it). Click on GO: Biological process to see an example of the GO pages.

Here you can see the functions that have been associated with the gene. There are three-letter codes that indicate how the association was made, as well as links to the specific transcript they are linked to.
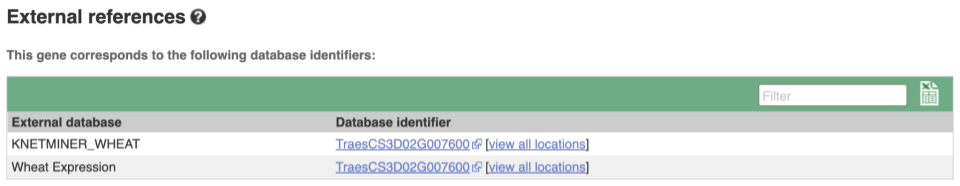
Gene information in external databases
We also have links out to other databases which have information about our genes and may focus on other topics that we don’t cover, like Expression Atlas or UniProtKB. Go up the left-hand menu to External references:
The transcript tab
We’re now going to explore the different transcripts of TraesCS3D02G007600. Click on Show transcript table at the top.
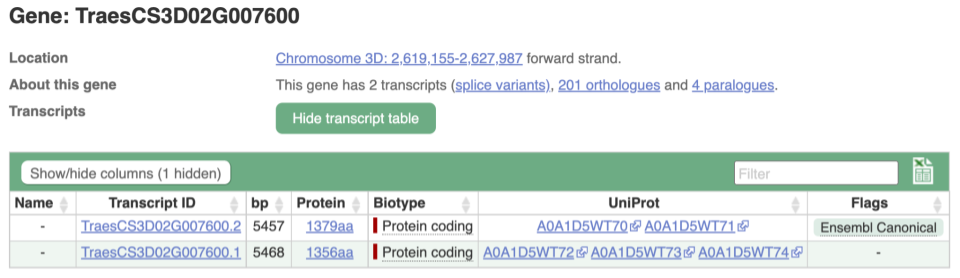
Here we can see a list of all the transcripts of TraesCS3D02G007600 with their identifiers, lengths and biotypes. Click on the ID of the Ensembl Canonical transcript, TraesCS3D02G007600.2.
You are now in the Transcript tab for TraesCS3D02G007600.2. We can still see the gene tab so we can easily jump back. The left hand navigation column provides several options for the transcript TraesCS3D02G007600.2 - many of these are similar to the options you see in the gene tab, but not all of them. If you can’t find the thing you’re looking for, often the solution is to switch tabs.
Transcript sequences
Click on the Exons link. This page is useful for designing RT-PCR primers because you can see the sequences of the different exons and their lengths.
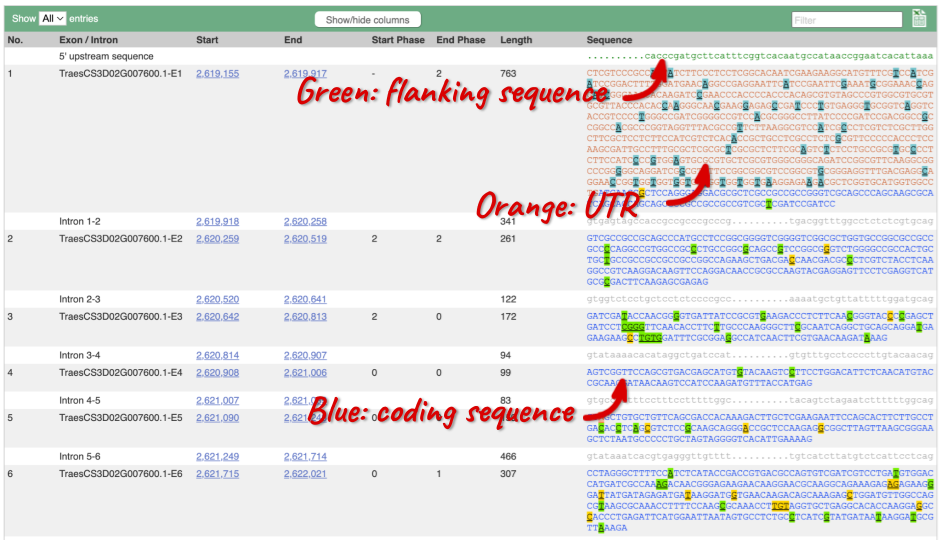
You may want to change the display (for example, to show more flanking sequence, or to show full introns). In order to do so click on Configure this page and change the display options accordingly.
Now click on the cDNA link to see the spliced transcript sequence with the amino acid sequence. This page is useful for mapping between the RNA and protein sequences, particularly genetic variants.
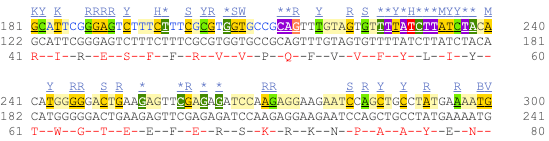
UnTranslated Regions (UTRs) are highlighted in dark yellow, codons are highlighted in light yellow, and exon sequence is shown in black or blue letters to show exon divides. Sequence variants are represented by highlighted nucleotides and clickable IUPAC codes are above the sequence.
Transcript information in external databases
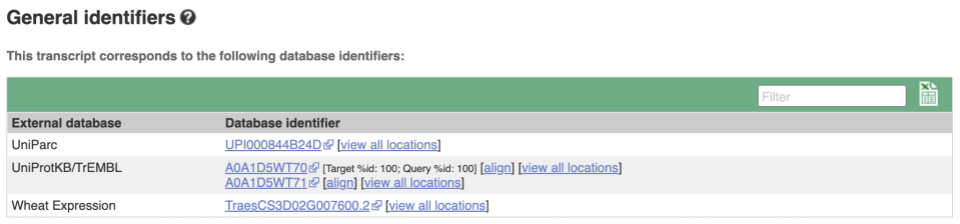
Next, follow the General identifiers link at the left. Just like the External References page in the gene tab, this page shows links out to other databases such as RefSeq, UniProtKB, PDBe and others, this time linked to the transcript or protein product, rather than the gene.
Protein domain information
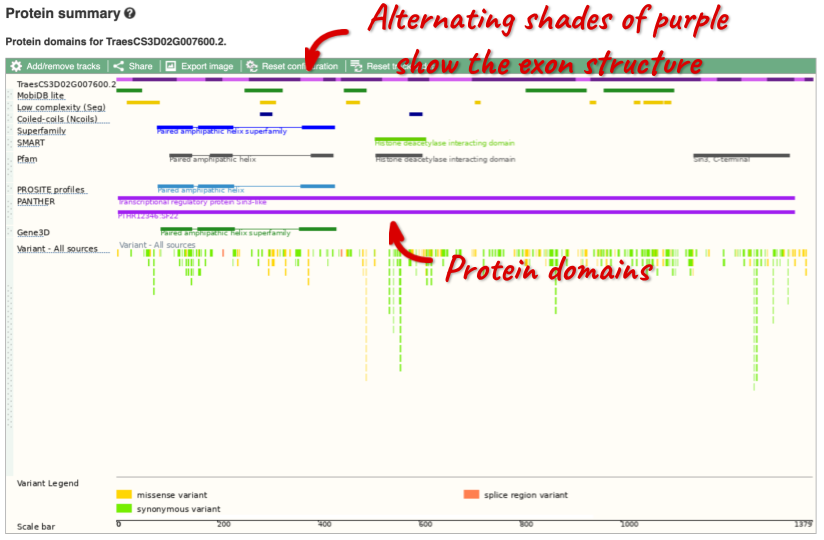
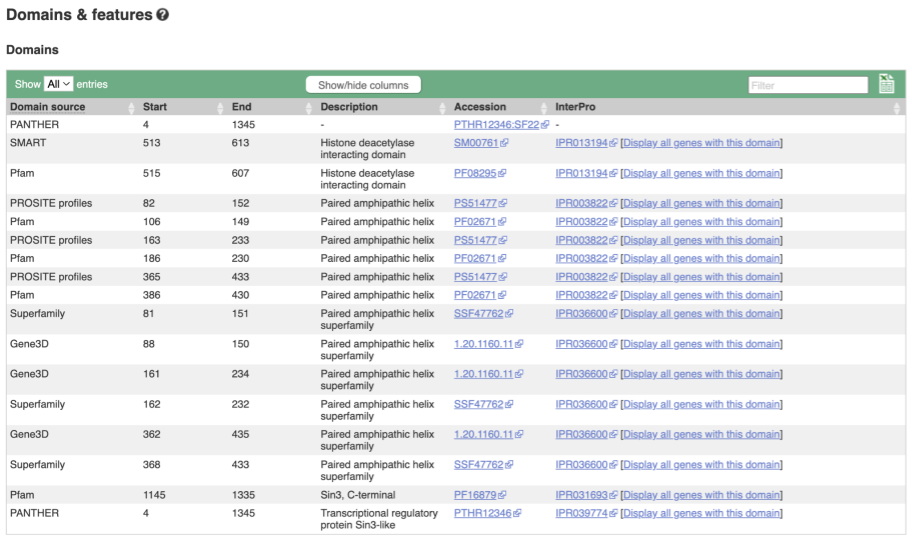
If you’re interested in protein domains, you could click on Protein summary to view domains from Pfam, PROSITE, Superfamily, InterPro, and more. These are all plotted against the transcript sequence, with the exons shown in alternating shades of purple at the top of the page. Alternatively, you can go to Domains & features to see a table of the same information.
Finding a Triticum aestivum gene
-
Search for
Oxygen evolving enhancer proteinfrom the Ensembl Plants homepage and narrow down your search to Triticum aestivum. How many genes are there with this name in wheat? Why do you think this is? What chromosomes are they on? -
Go to the gene on chromosome 2B. How many protein-coding transcripts does this gene have? What is a “canonical transcript”?
-
Click on the canonical transcript. How many exons does this transcript have? Export the protein sequence of this transcript in the FASTA format.
- Start at the Ensembl Plants homepage. Choose Triticum aestivum from the species drop-down, type
Oxygen evolving enhancer proteininto the search box then click Go.There are two genes named TraesCS2D02G248400 and TraesCS2B02G270300. This is because of the hybridisations in wheat’s evolutionary history. You can see that the two genes occur on chromosomes 2B and 2D.
- Click on the gene on chromosome 2B to go to the Gene tab. If the transcript table is hidden, click on Show transcript table to see it.
There are 2 protein coding transcripts.
Mouse over the Ensembl Canonical flag in the transcripts table to find a description.
The Ensembl canonical transcript is a single transcript chosen for each gene in each species. It is the most highly conserved, most highly expressed, has the longest coding sequence and is represented in other key resources (e.g. NCBI, UniProt)
- Click on TraesCS2B02G270300.2 in the transcript table. You can find the number of exons in the summary description at the top of the Summary page, or you can count the number of boxes (boxes represent exons, lines represent introns) in the Summary diagram.
TraesCS2B02G270300.2 has 2 exons.
Go to Sequence: Protein In the left-hand panel.
Click on the green Download sequence button above the protein sequence. Select FASTA from the drop-down in the pop-up menu and download the sequence to your local machine.
Exploring a Triticum aestivum (Wheat) gene
Start in the Ensembl Plants homepage and select the Triticum aestivum IWGSC genome to answer the following questions:
-
What GO: Molecular function terms are associated with the Wheat gene TraesCS6D02G180200?
-
Go to the transcript tab. How many exons does it have? Which one is the longest? Approximately, how much of that is coding?
-
What domains can be found in the protein product of this transcript? What prediction method(s) identified these domains?
- Go to Ensembl Plants, select Triticum aestivum from the drop down menu then type
TraesCS6D02G180200into the search box. Click on the gene name link TraesCS6D02G180200 in the search results. Click on GO: Molecular function in the left-hand menu.There is one term listed: GO:0005515, protein binding.
- Click on the transcript tab at the top of the page. Click on Exons in the left-hand menu.
There are six exons. Exon 6 is longest with 485 bp, of which around one sixth is coding.
- Click on either Protein Summary or Domains & features in the left-hand menu to view the data graphically or as a table, respectively.
Leucine-rich repeats are predicted by many different methods, however each method predict the leucine-rich repeats at different positions.







